Data Wrangling and Analyses with Tidyverse
Bracket subsetting is handy, but it can be cumbersome and difficult to read, especially for complicated operations.
Luckily, the dplyr
package provides a number of very useful functions for manipulating data
frames in a way that will reduce repetition, reduce the probability of
making errors, and probably even save you some typing. As an added
bonus, you might even find the dplyr grammar easier to
read.
Here we’re going to cover some of the most commonly used functions as
well as using pipes (%>%) to combine them:
glimpse()select()filter()group_by()summarize()mutate()pivot_longerandpivot_wider
Packages in R are sets of additional functions that let you do more
stuff in R. The functions we’ve been using, like str(),
come built into R; packages give you access to more functions. You need
to install a package and then load it to be able to use it.
install.packages("dplyr") ## installYou might get asked to choose a CRAN mirror – this is asking you to choose a site to download the package from. The choice doesn’t matter too much; I’d recommend choosing the RStudio mirror.
library("dplyr") ## loadYou only need to install a package once per computer, but you need to load it every time you open a new R session and want to use that package.
What is dplyr?
The package dplyr is a package that tries to provide
easy tools for the most common data manipulation tasks. This package is
also included in the tidyverse package,
which is a collection of eight different packages (dplyr,
ggplot2, tibble, tidyr,
readr, purrr, stringr, and
forcats). It is built to work directly with data frames.
The thinking behind it was largely inspired by the package
plyr which has been in use for some time but suffered from
being slow in some cases.dplyr addresses this by porting
much of the computation to C++. An additional feature is the ability to
work with data stored directly in an external database. The benefits of
doing this are that the data can be managed natively in a relational
database, queries can be conducted on that database, and only the
results of the query returned.
This addresses a common problem with R in that all operations are conducted in memory and thus the amount of data you can work with is limited by available memory. The database connections essentially remove that limitation in that you can have a database that is over 100s of GB, conduct queries on it directly and pull back just what you need for analysis in R.
Taking a quick look at data frames
Similar to str(), which comes built into R,
glimpse() is a dplyr function that (as the
name suggests) gives a glimpse of the data frame.
Rows: 1,704
Columns: 6
$ country <chr> "Afghanistan", "Afghanistan", "Afghanistan", "Afghanistan"…
$ year <int> 1952, 1957, 1962, 1967, 1972, 1977, 1982, 1987, 1992, 1997…
$ pop <dbl> 8425333, 9240934, 10267083, 11537966, 13079460, 14880372, …
$ continent <chr> "Asia", "Asia", "Asia", "Asia", "Asia", "Asia", "Asia", "A…
$ life_exp <dbl> 28.801, 30.332, 31.997, 34.020, 36.088, 38.438, 39.854, 40…
$ gdp_per_cap <dbl> 779.4453, 820.8530, 853.1007, 836.1971, 739.9811, 786.1134…In the above output, we can already gather some information about
gapminder, such as the number of rows and columns, column
names, type of vector in the columns, and the first few entries of each
column. Although what we see is similar to outputs of
str(), this method gives a cleaner visual output.
Note: What is a
tibble?In the
tidyversethere is a specialized version of adata.framecalled atibble. It largely behaves like adata.frame, but it has some considerate defaults when displaying its contents. We shall encounter some differences as we work through this section, and will highlight them as we go.
Coercing to a tibble
Let’s coerce our gapminder data.frame into
a tibble using the as_tibble() function. Let’s
first do this without assigning the result:
as_tibble(gapminder)# A tibble: 1,704 × 6
country year pop continent life_exp gdp_per_cap
<chr> <int> <dbl> <chr> <dbl> <dbl>
1 Afghanistan 1952 8425333 Asia 28.8 779.
2 Afghanistan 1957 9240934 Asia 30.3 821.
3 Afghanistan 1962 10267083 Asia 32.0 853.
4 Afghanistan 1967 11537966 Asia 34.0 836.
5 Afghanistan 1972 13079460 Asia 36.1 740.
6 Afghanistan 1977 14880372 Asia 38.4 786.
7 Afghanistan 1982 12881816 Asia 39.9 978.
8 Afghanistan 1987 13867957 Asia 40.8 852.
9 Afghanistan 1992 16317921 Asia 41.7 649.
10 Afghanistan 1997 22227415 Asia 41.8 635.
# … with 1,694 more rowsWhat do you notice?
- The preview helpfully tells us the dimensions at the top
# A tibble: 1,704 × 6. - The preview helpfully truncates the output as if we used
head(). - The preview tells us the
mode()of each of the columns.
To drive home the difference, do the following:
gapminderAnd notice that no such conveniences are afforded to us. So for the
remainder, we will coerce gapminder into a
tibble. However, we may interchangeably refer to a
tibble or a data.frame.
gapminder <- as_tibble(gapminder)Selecting columns and filtering rows
To select columns of a data frame, use select(). The
first argument to this function is the data frame
(gapminder), and the subsequent arguments are the columns
to keep.
select(gapminder, country, year, gdp_per_cap)# A tibble: 1,704 × 3
country year gdp_per_cap
<chr> <int> <dbl>
1 Afghanistan 1952 779.
2 Afghanistan 1957 821.
3 Afghanistan 1962 853.
4 Afghanistan 1967 836.
5 Afghanistan 1972 740.
6 Afghanistan 1977 786.
7 Afghanistan 1982 978.
8 Afghanistan 1987 852.
9 Afghanistan 1992 649.
10 Afghanistan 1997 635.
# … with 1,694 more rowsTo select all columns except certain ones, put a “-” in front of the variable to exclude it.
select(gapminder, -life_exp)# A tibble: 1,704 × 5
country year pop continent gdp_per_cap
<chr> <int> <dbl> <chr> <dbl>
1 Afghanistan 1952 8425333 Asia 779.
2 Afghanistan 1957 9240934 Asia 821.
3 Afghanistan 1962 10267083 Asia 853.
4 Afghanistan 1967 11537966 Asia 836.
5 Afghanistan 1972 13079460 Asia 740.
6 Afghanistan 1977 14880372 Asia 786.
7 Afghanistan 1982 12881816 Asia 978.
8 Afghanistan 1987 13867957 Asia 852.
9 Afghanistan 1992 16317921 Asia 649.
10 Afghanistan 1997 22227415 Asia 635.
# … with 1,694 more rowsdplyr also provides useful functions to select columns
based on their names. For instance, ends_with() allows you
to select columns that ends with specific letters. For instance, if you
wanted to select columns that end with the letter “p”:
select(gapminder, ends_with("p"))# A tibble: 1,704 × 3
pop life_exp gdp_per_cap
<dbl> <dbl> <dbl>
1 8425333 28.8 779.
2 9240934 30.3 821.
3 10267083 32.0 853.
4 11537966 34.0 836.
5 13079460 36.1 740.
6 14880372 38.4 786.
7 12881816 39.9 978.
8 13867957 40.8 852.
9 16317921 41.7 649.
10 22227415 41.8 635.
# … with 1,694 more rowsChallenge
Create a table that contains all the columns with the letter “e” and column “country”, without columns “life_exp”. Hint: look at the help function
tidyselect::ends_with()we’ve just covered.
Solution
select(gapminder, contains("e"), -life_exp, country)# A tibble: 1,704 × 4
year continent gdp_per_cap country
<int> <chr> <dbl> <chr>
1 1952 Asia 779. Afghanistan
2 1957 Asia 821. Afghanistan
3 1962 Asia 853. Afghanistan
4 1967 Asia 836. Afghanistan
5 1972 Asia 740. Afghanistan
6 1977 Asia 786. Afghanistan
7 1982 Asia 978. Afghanistan
8 1987 Asia 852. Afghanistan
9 1992 Asia 649. Afghanistan
10 1997 Asia 635. Afghanistan
# … with 1,694 more rowsTo choose rows, use filter():
filter(gapminder, country == 'Nigeria')# A tibble: 12 × 6
country year pop continent life_exp gdp_per_cap
<chr> <int> <dbl> <chr> <dbl> <dbl>
1 Nigeria 1952 33119096 Africa 36.3 1077.
2 Nigeria 1957 37173340 Africa 37.8 1101.
3 Nigeria 1962 41871351 Africa 39.4 1151.
4 Nigeria 1967 47287752 Africa 41.0 1015.
5 Nigeria 1972 53740085 Africa 42.8 1698.
6 Nigeria 1977 62209173 Africa 44.5 1982.
7 Nigeria 1982 73039376 Africa 45.8 1577.
8 Nigeria 1987 81551520 Africa 46.9 1385.
9 Nigeria 1992 93364244 Africa 47.5 1620.
10 Nigeria 1997 106207839 Africa 47.5 1625.
11 Nigeria 2002 119901274 Africa 46.6 1615.
12 Nigeria 2007 135031164 Africa 46.9 2014.filter() will keep all the rows that match the
conditions that are provided. Here are a few examples:
# rows for which the country column contains Vietnam or Indonesia
filter(gapminder, country %in% c('Vietnam', 'Indonesia'))# A tibble: 24 × 6
country year pop continent life_exp gdp_per_cap
<chr> <int> <dbl> <chr> <dbl> <dbl>
1 Indonesia 1952 82052000 Asia 37.5 750.
2 Indonesia 1957 90124000 Asia 39.9 859.
3 Indonesia 1962 99028000 Asia 42.5 849.
4 Indonesia 1967 109343000 Asia 46.0 762.
5 Indonesia 1972 121282000 Asia 49.2 1111.
6 Indonesia 1977 136725000 Asia 52.7 1383.
7 Indonesia 1982 153343000 Asia 56.2 1517.
8 Indonesia 1987 169276000 Asia 60.1 1748.
9 Indonesia 1992 184816000 Asia 62.7 2383.
10 Indonesia 1997 199278000 Asia 66.0 3119.
# … with 14 more rows# rows with life_exp greater than or equal to 70
filter(gapminder, life_exp >= 70)# A tibble: 494 × 6
country year pop continent life_exp gdp_per_cap
<chr> <int> <dbl> <chr> <dbl> <dbl>
1 Albania 1982 2780097 Europe 70.4 3631.
2 Albania 1987 3075321 Europe 72 3739.
3 Albania 1992 3326498 Europe 71.6 2497.
4 Albania 1997 3428038 Europe 73.0 3193.
5 Albania 2002 3508512 Europe 75.7 4604.
6 Albania 2007 3600523 Europe 76.4 5937.
7 Algeria 2002 31287142 Africa 71.0 5288.
8 Algeria 2007 33333216 Africa 72.3 6223.
9 Argentina 1987 31620918 Americas 70.8 9140.
10 Argentina 1992 33958947 Americas 71.9 9308.
# … with 484 more rowsfilter() allows you to combine multiple conditions. You
can separate them using a , as arguments to the function,
they will be combined using the & (AND) logical
operator. If you need to use the | (OR) logical operator,
you can specify it explicitly:
# this is equivalent to:
# filter(gapminder, country == "Germany" & year >= 1980)
filter(gapminder, country == "Germany", year >= 1980)# A tibble: 6 × 6
country year pop continent life_exp gdp_per_cap
<chr> <int> <dbl> <chr> <dbl> <dbl>
1 Germany 1982 78335266 Europe 73.8 22032.
2 Germany 1987 77718298 Europe 74.8 24639.
3 Germany 1992 80597764 Europe 76.1 26505.
4 Germany 1997 82011073 Europe 77.3 27789.
5 Germany 2002 82350671 Europe 78.7 30036.
6 Germany 2007 82400996 Europe 79.4 32170.# using `|` logical operator
filter(gapminder, year >= 1990, (country == "Australia" | country == 'Mauritius'))# A tibble: 8 × 6
country year pop continent life_exp gdp_per_cap
<chr> <int> <dbl> <chr> <dbl> <dbl>
1 Australia 1992 17481977 Oceania 77.6 23425.
2 Australia 1997 18565243 Oceania 78.8 26998.
3 Australia 2002 19546792 Oceania 80.4 30688.
4 Australia 2007 20434176 Oceania 81.2 34435.
5 Mauritius 1992 1096202 Africa 69.7 6058.
6 Mauritius 1997 1149818 Africa 70.7 7426.
7 Mauritius 2002 1200206 Africa 72.0 9022.
8 Mauritius 2007 1250882 Africa 72.8 10957.Challenge
Select all data for countries in Europe between the years 1990 and 2000
Solution
filter(gapminder, continent == 'Europe', (year >= 1990 & year <= 2000))# A tibble: 60 × 6
country year pop continent life_exp gdp_per_cap
<chr> <int> <dbl> <chr> <dbl> <dbl>
1 Albania 1992 3326498 Europe 71.6 2497.
2 Albania 1997 3428038 Europe 73.0 3193.
3 Austria 1992 7914969 Europe 76.0 27042.
4 Austria 1997 8069876 Europe 77.5 29096.
5 Belgium 1992 10045622 Europe 76.5 25576.
6 Belgium 1997 10199787 Europe 77.5 27561.
7 Bosnia and Herzegovina 1992 4256013 Europe 72.2 2547.
8 Bosnia and Herzegovina 1997 3607000 Europe 73.2 4766.
9 Bulgaria 1992 8658506 Europe 71.2 6303.
10 Bulgaria 1997 8066057 Europe 70.3 5970.
# … with 50 more rowsPipes
But what if you wanted to select and filter? We can do this with
pipes (which we saw in the bash episode). Pipes, are a
fairly recent addition to R. Pipes let you take the output of one
function and send it directly to the next, which is useful when you need
to many things to the same data set. Pipes in R look like
%>% (recall they looked like | in
bash) and are available via the magrittr
package, which is installed as part of dplyr. If you use
RStudio, you can type the pipe with Ctrl + Shift +
M if you’re using a PC, or Cmd + Shift
+ M if you’re using a Mac.
gapminder %>%
filter(country == "Spain") %>%
select(year, pop, life_exp)# A tibble: 12 × 3
year pop life_exp
<int> <dbl> <dbl>
1 1952 28549870 64.9
2 1957 29841614 66.7
3 1962 31158061 69.7
4 1967 32850275 71.4
5 1972 34513161 73.1
6 1977 36439000 74.4
7 1982 37983310 76.3
8 1987 38880702 76.9
9 1992 39549438 77.6
10 1997 39855442 78.8
11 2002 40152517 79.8
12 2007 40448191 80.9In the above code, we use the pipe to send the gapminder
dataset first through filter(), to keep rows where
country matches a particular country, and then through
select() to keep only the year,
pop, and life_exp columns. Since
%>% takes the object on its left and passes it as the
first argument to the function on its right, we don’t need to explicitly
include the data frame as an argument to the filter() and
select() functions any more.
Some may find it helpful to read the pipe like the word “then”. For
instance, in the above example, we took the data frame
gapminder, then we filtered for rows
where country was Spain, then we
selected the year, pop, and
life_exp columns, then we showed only the first
six rows. The dplyr functions by
themselves are somewhat simple, but by combining them into linear
workflows with the pipe, we can accomplish more complex manipulations of
data frames.
If we want to create a new object with this smaller version of the data we can do so by assigning it a new name:
spain_gapminder <- gapminder %>%
filter(country == "Spain") %>%
select(year, pop, life_exp)This new object includes all of the data from this sample. Let’s look at just the first six rows to confirm it’s what we want:
spain_gapminder# A tibble: 12 × 3
year pop life_exp
<int> <dbl> <dbl>
1 1952 28549870 64.9
2 1957 29841614 66.7
3 1962 31158061 69.7
4 1967 32850275 71.4
5 1972 34513161 73.1
6 1977 36439000 74.4
7 1982 37983310 76.3
8 1987 38880702 76.9
9 1992 39549438 77.6
10 1997 39855442 78.8
11 2002 40152517 79.8
12 2007 40448191 80.9Similar to head() and tail() functions, we
can also look at the first or last six rows using tidyverse function
slice(). Slice is a more versatile function that allows
users to specify a range to view:
spain_gapminder %>% slice(1:6)# A tibble: 6 × 3
year pop life_exp
<int> <dbl> <dbl>
1 1952 28549870 64.9
2 1957 29841614 66.7
3 1962 31158061 69.7
4 1967 32850275 71.4
5 1972 34513161 73.1
6 1977 36439000 74.4spain_gapminder %>% slice(7:11)# A tibble: 5 × 3
year pop life_exp
<int> <dbl> <dbl>
1 1982 37983310 76.3
2 1987 38880702 76.9
3 1992 39549438 77.6
4 1997 39855442 78.8
5 2002 40152517 79.8Exercise: Pipe and filter
Starting with the
gapminderdata frame, use pipes to subset the data to include only observations from Panama, where the year is at least 1980. Showing only the 4th through 6th rows of columnscountry,year, andgdp_per_cap.
Solution
gapminder %>%
filter(country == "Panama" & year >= 1980) %>%
slice(4:6) %>%
select(country, year, gdp_per_cap)# A tibble: 3 × 3
country year gdp_per_cap
<chr> <int> <dbl>
1 Panama 1997 7114.
2 Panama 2002 7356.
3 Panama 2007 9809.Mutate
Frequently you’ll want to create new columns based on the values in
existing columns, for example to do unit conversions or find the ratio
of values in two columns. For this we’ll use the dplyr
function mutate().
We have a column titled “gdp_per_cap” and “pop”. We could use these two columns to compute the “total_gdp” for each country/year observation. By multiplying the entries per-row.
Let’s add a column (total_gdp) to our
gapminder data frame that shows the total GDP for the
country in the corresponding year.
gapminder %>% mutate(total_gdp = gdp_per_cap * pop)# A tibble: 1,704 × 7
country year pop continent life_exp gdp_per_cap total_gdp
<chr> <int> <dbl> <chr> <dbl> <dbl> <dbl>
1 Afghanistan 1952 8425333 Asia 28.8 779. 6567086330.
2 Afghanistan 1957 9240934 Asia 30.3 821. 7585448670.
3 Afghanistan 1962 10267083 Asia 32.0 853. 8758855797.
4 Afghanistan 1967 11537966 Asia 34.0 836. 9648014150.
5 Afghanistan 1972 13079460 Asia 36.1 740. 9678553274.
6 Afghanistan 1977 14880372 Asia 38.4 786. 11697659231.
7 Afghanistan 1982 12881816 Asia 39.9 978. 12598563401.
8 Afghanistan 1987 13867957 Asia 40.8 852. 11820990309.
9 Afghanistan 1992 16317921 Asia 41.7 649. 10595901589.
10 Afghanistan 1997 22227415 Asia 41.8 635. 14121995875.
# … with 1,694 more rowsExercise
There is data for a lot of countries and years, so let’s look just at the results of the United States by adding the correct line to the above code.
Solution
gapminder %>%
mutate(total_gdp = gdp_per_cap * pop) %>%
filter(country == 'United States')# A tibble: 12 × 7
country year pop continent life_exp gdp_per_cap total_gdp
<chr> <int> <dbl> <chr> <dbl> <dbl> <dbl>
1 United States 1952 157553000 Americas 68.4 13990. 2.20e12
2 United States 1957 171984000 Americas 69.5 14847. 2.55e12
3 United States 1962 186538000 Americas 70.2 16173. 3.02e12
4 United States 1967 198712000 Americas 70.8 19530. 3.88e12
5 United States 1972 209896000 Americas 71.3 21806. 4.58e12
6 United States 1977 220239000 Americas 73.4 24073. 5.30e12
7 United States 1982 232187835 Americas 74.6 25010. 5.81e12
8 United States 1987 242803533 Americas 75.0 29884. 7.26e12
9 United States 1992 256894189 Americas 76.1 32004. 8.22e12
10 United States 1997 272911760 Americas 76.8 35767. 9.76e12
11 United States 2002 287675526 Americas 77.3 39097. 1.12e13
12 United States 2007 301139947 Americas 78.2 42952. 1.29e13group_by() and summarize() functions
Many data analysis tasks can be approached using the
“split-apply-combine” paradigm: split the data into groups, apply some
analysis to each group, and then combine the results. dplyr
makes this very easy through the use of the group_by()
function, which splits the data into groups. When the data is grouped in
this way summarize() can be used to collapse each group
into a single-row summary. summarize() does this by
applying an aggregating or summary function to each group. For example,
if we wanted to group by continent and find the number of rows of data
for each continent, we would do:
gapminder %>%
group_by(continent) %>%
summarize(n())# A tibble: 5 × 2
continent `n()`
<chr> <int>
1 Africa 624
2 Americas 300
3 Asia 396
4 Europe 360
5 Oceania 24Notice this is the same results as when we ran
summary(gapminder$continent).
It can be a bit tricky at first, but we can imagine splitting the data frame by groups and applying a certain function to summarize the data.
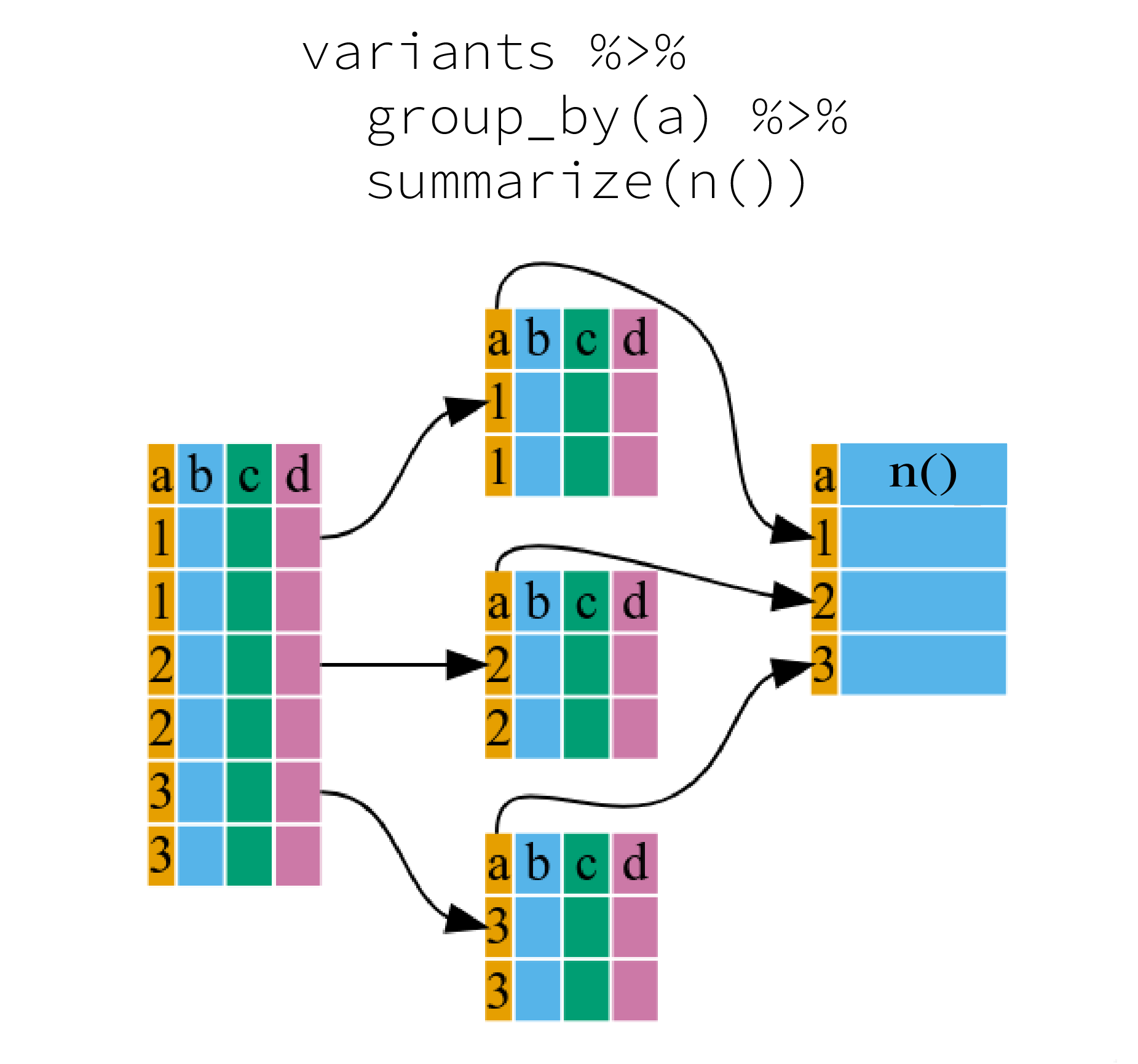
Here the summary function used was n() to find the count
for each group. Since this is a quite a common operation, there is a
simpler method called tally():
gapminder %>%
group_by(country) %>%
tally()# A tibble: 142 × 2
country n
<chr> <int>
1 Afghanistan 12
2 Albania 12
3 Algeria 12
4 Angola 12
5 Argentina 12
6 Australia 12
7 Austria 12
8 Bahrain 12
9 Bangladesh 12
10 Belgium 12
# … with 132 more rowsTo show that there are many ways to achieve the same results, there
is another way to approach this, which bypasses group_by()
using the function count():
gapminder %>% count(country)# A tibble: 142 × 2
country n
<chr> <int>
1 Afghanistan 12
2 Albania 12
3 Algeria 12
4 Angola 12
5 Argentina 12
6 Australia 12
7 Austria 12
8 Bahrain 12
9 Bangladesh 12
10 Belgium 12
# … with 132 more rowsWe can also apply many other functions to individual columns to get
other summary statistics. For example,we can use built-in functions like
mean(), median(), min(), and
max(). These are called “built-in functions” because they
come with R and don’t require that you install any additional packages.
By default, all R functions operating on vectors that contains
missing data will return NA. It’s a way to make sure that users
know they have missing data, and make a conscious decision on how to
deal with it. When dealing with simple statistics like the mean, the
easiest way to ignore NA (the missing data) is to use
na.rm = TRUE (rm stands for remove).
So to view the mean, median, maximum, and minimum
gdp_per_cap for each country:
gapminder %>%
group_by(country) %>%
summarize(
min_gpc = min(gdp_per_cap),
mean_gpc = mean(gdp_per_cap),
median_gpc = median(gdp_per_cap),
max_gpc = max(gdp_per_cap))# A tibble: 142 × 5
country min_gpc mean_gpc median_gpc max_gpc
<chr> <dbl> <dbl> <dbl> <dbl>
1 Afghanistan 635. 803. 803. 978.
2 Albania 1601. 3255. 3253. 5937.
3 Algeria 2449. 4426. 4854. 6223.
4 Angola 2277. 3607. 3265. 5523.
5 Argentina 5911. 8956. 9069. 12779.
6 Australia 10040. 19981. 18906. 34435.
7 Austria 6137. 20412. 20673. 36126.
8 Bahrain 9867. 18078. 18780. 29796.
9 Bangladesh 630. 818. 704. 1391.
10 Belgium 8343. 19901. 20049. 33693.
# … with 132 more rowsReshaping data frames
It can sometimes be useful to transform the “long” tidy format, into
the wide format. This transformation can be done with the
pivot_wider() function provided by the tidyr
package (also part of the tidyverse).
pivot_wider() takes a data frame as the first argument,
and two arguments: the column name that will become the columns and the
column name that will become the cells in the wide data. Let’s create a
wide format table with rows for each country, columns for each year, and
data values being the life_exp.
gapminder_wide <- gapminder %>%
select(country, life_exp, year) %>%
group_by(country) %>%
pivot_wider(names_from = year, values_from = life_exp)
gapminder_wide# A tibble: 142 × 13
# Groups: country [142]
country `1952` `1957` `1962` `1967` `1972` `1977` `1982` `1987` `1992` `1997`
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Afghan… 28.8 30.3 32.0 34.0 36.1 38.4 39.9 40.8 41.7 41.8
2 Albania 55.2 59.3 64.8 66.2 67.7 68.9 70.4 72 71.6 73.0
3 Algeria 43.1 45.7 48.3 51.4 54.5 58.0 61.4 65.8 67.7 69.2
4 Angola 30.0 32.0 34 36.0 37.9 39.5 39.9 39.9 40.6 41.0
5 Argent… 62.5 64.4 65.1 65.6 67.1 68.5 69.9 70.8 71.9 73.3
6 Austra… 69.1 70.3 70.9 71.1 71.9 73.5 74.7 76.3 77.6 78.8
7 Austria 66.8 67.5 69.5 70.1 70.6 72.2 73.2 74.9 76.0 77.5
8 Bahrain 50.9 53.8 56.9 59.9 63.3 65.6 69.1 70.8 72.6 73.9
9 Bangla… 37.5 39.3 41.2 43.5 45.3 46.9 50.0 52.8 56.0 59.4
10 Belgium 68 69.2 70.2 70.9 71.4 72.8 73.9 75.4 76.5 77.5
# … with 132 more rows, and 2 more variables: 2002 <dbl>, 2007 <dbl>The opposite operation of pivot_wider() is taken care by
pivot_longer(). We specify the names of the new columns,
and here add -CHROM as this column shouldn’t be affected by
the reshaping:
gapminder_wide %>%
pivot_longer(-country, names_to = "year", values_to = "life_exp")# A tibble: 1,704 × 3
# Groups: country [142]
country year life_exp
<chr> <chr> <dbl>
1 Afghanistan 1952 28.8
2 Afghanistan 1957 30.3
3 Afghanistan 1962 32.0
4 Afghanistan 1967 34.0
5 Afghanistan 1972 36.1
6 Afghanistan 1977 38.4
7 Afghanistan 1982 39.9
8 Afghanistan 1987 40.8
9 Afghanistan 1992 41.7
10 Afghanistan 1997 41.8
# … with 1,694 more rowsThe figure was adapted from the Software Carpentry lesson, R for Reproducible Scientific Analysis↩︎