Data Wrangling and Analyses with Tidyverse
Overview
The R multi-verse
In the remaining lessons we will focus on manipulating data with
packages that are part of the tidyverse, namely
dplyr, tidyr, and ggplot2. The
tidyverse describes itself as “an opinionated collection of R packages
designed for data science. All packages share an underlying design
philosophy, grammar, and data structures.” We have already spoken about
the idea of “base R,” which is a set of built-in functions that have an
implied coding style. Indeed, the lessons up until now have not required
us to load any packages.
In particular, dplyr and tidyr, along with
the companion package tibble, are a “modern re-imagnining”
of the data frame and its manipulation. The functions that accomplish
many common data manipulation challenges are clearly named, and can be
linked together in a way that makes code simple and readable, as we
shall see. Of course, as with many programming languages, there are many
ways to accomplish the same task, and nearly every
tidyverse way of doing something can be translated into
base R and vice-versa.
A question many people have when they’re learning R is, “Why learn
base R ways of doing things if I can use the easy tidyverse
way?” If you’re dealing primarily with data in data frames, you can
probably get by with ignoring base R ways of doing things. And, truth be
told, many people will only deal in data frames.
However, if you are analyzing data in a
bioinformatics context, you are likely going to be working with
Bioconductor packages, and these packages are not necessarily designed
to work with the tidyverse because the data structures
involved are more complex than simple data frames (as we will see in
RNA-seq Demystified). Consequently, knowing about R objects other than
data frames, and knowing how to access, subset, and otherwise manipulate
those objects, will make working with Bioconductor packages easier.
So rather than either/or, we should think of it as both/and. Knowing
base R and its way of doing things will help you in some contexts, while
knowing tidyverse will help you in others.
In this lesson
We’re going to cover some of the most commonly used functions as well
as tibbles to store data and pipes (%>%) to combine
functions:
glimpse()select()filter()group_by()summarize()mutate()pivot_longerpivot_wider
Functions for data
To get started, let’s load the tidyverse package:
library(tidyverse)Tibbles
While the dplyr packages operate on data frames with
aplomb, the tidyverse tends to use a tibble, which it
describes as “a modern re-imagining of the data frame.” What are some of
the benefits of a tibble over a data frame?
- It never changes input types. Remember
stringsAsFactors? A tibble never converts strings to factors. - It never changes variable names. If you try to create a
data.framewith column names beginning with numbers or containing dashes, those characters will be replaced. A tibble will use what you give it. - It has a humane
print()function that won’t try to show you thousands of rows of data.
Let’s coerce our gapminder data frame into a tibble
using the as_tibble() function. Let’s first do this without
assigning the result:
as_tibble(gapminder)# A tibble: 1,704 × 6
country year pop continent lifeExp gdpPercap
<chr> <int> <dbl> <chr> <dbl> <dbl>
1 Afghanistan 1952 8425333 Asia 28.8 779.
2 Afghanistan 1957 9240934 Asia 30.3 821.
3 Afghanistan 1962 10267083 Asia 32.0 853.
4 Afghanistan 1967 11537966 Asia 34.0 836.
5 Afghanistan 1972 13079460 Asia 36.1 740.
6 Afghanistan 1977 14880372 Asia 38.4 786.
7 Afghanistan 1982 12881816 Asia 39.9 978.
8 Afghanistan 1987 13867957 Asia 40.8 852.
9 Afghanistan 1992 16317921 Asia 41.7 649.
10 Afghanistan 1997 22227415 Asia 41.8 635.
# … with 1,694 more rowsNotice that the print function basically shows us a preview of the table, its dimensions, and the mode of each column. That’s nice and helpful!
Now let’s print gapminder in its data frame form:
gapminder country year pop continent lifeExp gdpPercap
1 Afghanistan 1952 8425333 Asia 28.801 779.4453
2 Afghanistan 1957 9240934 Asia 30.332 820.8530
3 Afghanistan 1962 10267083 Asia 31.997 853.1007
4 Afghanistan 1967 11537966 Asia 34.020 836.1971
5 Afghanistan 1972 13079460 Asia 36.088 739.9811
6 Afghanistan 1977 14880372 Asia 38.438 786.1134
7 Afghanistan 1982 12881816 Asia 39.854 978.0114
8 Afghanistan 1987 13867957 Asia 40.822 852.3959
9 Afghanistan 1992 16317921 Asia 41.674 649.3414
10 Afghanistan 1997 22227415 Asia 41.763 635.3414
11 Afghanistan 2002 25268405 Asia 42.129 726.7341
12 Afghanistan 2007 31889923 Asia 43.828 974.5803
13 Albania 1952 1282697 Europe 55.230 1601.0561
14 Albania 1957 1476505 Europe 59.280 1942.2842
15 Albania 1962 1728137 Europe 64.820 2312.8890
16 Albania 1967 1984060 Europe 66.220 2760.1969
17 Albania 1972 2263554 Europe 67.690 3313.4222
18 Albania 1977 2509048 Europe 68.930 3533.0039
19 Albania 1982 2780097 Europe 70.420 3630.8807
20 Albania 1987 3075321 Europe 72.000 3738.9327
21 Albania 1992 3326498 Europe 71.581 2497.4379
22 Albania 1997 3428038 Europe 72.950 3193.0546
23 Albania 2002 3508512 Europe 75.651 4604.2117
24 Albania 2007 3600523 Europe 76.423 5937.0295
25 Algeria 1952 9279525 Africa 43.077 2449.0082
26 Algeria 1957 10270856 Africa 45.685 3013.9760
27 Algeria 1962 11000948 Africa 48.303 2550.8169
28 Algeria 1967 12760499 Africa 51.407 3246.9918
29 Algeria 1972 14760787 Africa 54.518 4182.6638
30 Algeria 1977 17152804 Africa 58.014 4910.4168
31 Algeria 1982 20033753 Africa 61.368 5745.1602
32 Algeria 1987 23254956 Africa 65.799 5681.3585
33 Algeria 1992 26298373 Africa 67.744 5023.2166
34 Algeria 1997 29072015 Africa 69.152 4797.2951
35 Algeria 2002 31287142 Africa 70.994 5288.0404
36 Algeria 2007 33333216 Africa 72.301 6223.3675
37 Angola 1952 4232095 Africa 30.015 3520.6103
38 Angola 1957 4561361 Africa 31.999 3827.9405
39 Angola 1962 4826015 Africa 34.000 4269.2767
40 Angola 1967 5247469 Africa 35.985 5522.7764
41 Angola 1972 5894858 Africa 37.928 5473.2880
42 Angola 1977 6162675 Africa 39.483 3008.6474
43 Angola 1982 7016384 Africa 39.942 2756.9537
44 Angola 1987 7874230 Africa 39.906 2430.2083
45 Angola 1992 8735988 Africa 40.647 2627.8457
46 Angola 1997 9875024 Africa 40.963 2277.1409
47 Angola 2002 10866106 Africa 41.003 2773.2873
48 Angola 2007 12420476 Africa 42.731 4797.2313
49 Argentina 1952 17876956 Americas 62.485 5911.3151
50 Argentina 1957 19610538 Americas 64.399 6856.8562
51 Argentina 1962 21283783 Americas 65.142 7133.1660
52 Argentina 1967 22934225 Americas 65.634 8052.9530
53 Argentina 1972 24779799 Americas 67.065 9443.0385
54 Argentina 1977 26983828 Americas 68.481 10079.0267
55 Argentina 1982 29341374 Americas 69.942 8997.8974
56 Argentina 1987 31620918 Americas 70.774 9139.6714
57 Argentina 1992 33958947 Americas 71.868 9308.4187
58 Argentina 1997 36203463 Americas 73.275 10967.2820
59 Argentina 2002 38331121 Americas 74.340 8797.6407
60 Argentina 2007 40301927 Americas 75.320 12779.3796
61 Australia 1952 8691212 Oceania 69.120 10039.5956
62 Australia 1957 9712569 Oceania 70.330 10949.6496
63 Australia 1962 10794968 Oceania 70.930 12217.2269
64 Australia 1967 11872264 Oceania 71.100 14526.1246
65 Australia 1972 13177000 Oceania 71.930 16788.6295
66 Australia 1977 14074100 Oceania 73.490 18334.1975
67 Australia 1982 15184200 Oceania 74.740 19477.0093
68 Australia 1987 16257249 Oceania 76.320 21888.8890
69 Australia 1992 17481977 Oceania 77.560 23424.7668
70 Australia 1997 18565243 Oceania 78.830 26997.9366
71 Australia 2002 19546792 Oceania 80.370 30687.7547
72 Australia 2007 20434176 Oceania 81.235 34435.3674
73 Austria 1952 6927772 Europe 66.800 6137.0765
74 Austria 1957 6965860 Europe 67.480 8842.5980
75 Austria 1962 7129864 Europe 69.540 10750.7211
76 Austria 1967 7376998 Europe 70.140 12834.6024
77 Austria 1972 7544201 Europe 70.630 16661.6256
78 Austria 1977 7568430 Europe 72.170 19749.4223
79 Austria 1982 7574613 Europe 73.180 21597.0836
80 Austria 1987 7578903 Europe 74.940 23687.8261
81 Austria 1992 7914969 Europe 76.040 27042.0187
82 Austria 1997 8069876 Europe 77.510 29095.9207
83 Austria 2002 8148312 Europe 78.980 32417.6077
84 Austria 2007 8199783 Europe 79.829 36126.4927
85 Bahrain 1952 120447 Asia 50.939 9867.0848
86 Bahrain 1957 138655 Asia 53.832 11635.7995
87 Bahrain 1962 171863 Asia 56.923 12753.2751
88 Bahrain 1967 202182 Asia 59.923 14804.6727
89 Bahrain 1972 230800 Asia 63.300 18268.6584
90 Bahrain 1977 297410 Asia 65.593 19340.1020
91 Bahrain 1982 377967 Asia 69.052 19211.1473
92 Bahrain 1987 454612 Asia 70.750 18524.0241
93 Bahrain 1992 529491 Asia 72.601 19035.5792
94 Bahrain 1997 598561 Asia 73.925 20292.0168
95 Bahrain 2002 656397 Asia 74.795 23403.5593
96 Bahrain 2007 708573 Asia 75.635 29796.0483
97 Bangladesh 1952 46886859 Asia 37.484 684.2442
98 Bangladesh 1957 51365468 Asia 39.348 661.6375
99 Bangladesh 1962 56839289 Asia 41.216 686.3416
100 Bangladesh 1967 62821884 Asia 43.453 721.1861
101 Bangladesh 1972 70759295 Asia 45.252 630.2336
102 Bangladesh 1977 80428306 Asia 46.923 659.8772
103 Bangladesh 1982 93074406 Asia 50.009 676.9819
104 Bangladesh 1987 103764241 Asia 52.819 751.9794
105 Bangladesh 1992 113704579 Asia 56.018 837.8102
106 Bangladesh 1997 123315288 Asia 59.412 972.7700
107 Bangladesh 2002 135656790 Asia 62.013 1136.3904
108 Bangladesh 2007 150448339 Asia 64.062 1391.2538
109 Belgium 1952 8730405 Europe 68.000 8343.1051
110 Belgium 1957 8989111 Europe 69.240 9714.9606
111 Belgium 1962 9218400 Europe 70.250 10991.2068
112 Belgium 1967 9556500 Europe 70.940 13149.0412
113 Belgium 1972 9709100 Europe 71.440 16672.1436
114 Belgium 1977 9821800 Europe 72.800 19117.9745
115 Belgium 1982 9856303 Europe 73.930 20979.8459
116 Belgium 1987 9870200 Europe 75.350 22525.5631
117 Belgium 1992 10045622 Europe 76.460 25575.5707
118 Belgium 1997 10199787 Europe 77.530 27561.1966
119 Belgium 2002 10311970 Europe 78.320 30485.8838
120 Belgium 2007 10392226 Europe 79.441 33692.6051
121 Benin 1952 1738315 Africa 38.223 1062.7522
122 Benin 1957 1925173 Africa 40.358 959.6011
123 Benin 1962 2151895 Africa 42.618 949.4991
124 Benin 1967 2427334 Africa 44.885 1035.8314
125 Benin 1972 2761407 Africa 47.014 1085.7969
126 Benin 1977 3168267 Africa 49.190 1029.1613
127 Benin 1982 3641603 Africa 50.904 1277.8976
128 Benin 1987 4243788 Africa 52.337 1225.8560
129 Benin 1992 4981671 Africa 53.919 1191.2077
130 Benin 1997 6066080 Africa 54.777 1232.9753
131 Benin 2002 7026113 Africa 54.406 1372.8779
132 Benin 2007 8078314 Africa 56.728 1441.2849
133 Bolivia 1952 2883315 Americas 40.414 2677.3263
134 Bolivia 1957 3211738 Americas 41.890 2127.6863
135 Bolivia 1962 3593918 Americas 43.428 2180.9725
136 Bolivia 1967 4040665 Americas 45.032 2586.8861
137 Bolivia 1972 4565872 Americas 46.714 2980.3313
138 Bolivia 1977 5079716 Americas 50.023 3548.0978
139 Bolivia 1982 5642224 Americas 53.859 3156.5105
140 Bolivia 1987 6156369 Americas 57.251 2753.6915
141 Bolivia 1992 6893451 Americas 59.957 2961.6997
142 Bolivia 1997 7693188 Americas 62.050 3326.1432
143 Bolivia 2002 8445134 Americas 63.883 3413.2627
144 Bolivia 2007 9119152 Americas 65.554 3822.1371
145 Bosnia and Herzegovina 1952 2791000 Europe 53.820 973.5332
146 Bosnia and Herzegovina 1957 3076000 Europe 58.450 1353.9892
147 Bosnia and Herzegovina 1962 3349000 Europe 61.930 1709.6837
148 Bosnia and Herzegovina 1967 3585000 Europe 64.790 2172.3524
149 Bosnia and Herzegovina 1972 3819000 Europe 67.450 2860.1698
150 Bosnia and Herzegovina 1977 4086000 Europe 69.860 3528.4813
151 Bosnia and Herzegovina 1982 4172693 Europe 70.690 4126.6132
152 Bosnia and Herzegovina 1987 4338977 Europe 71.140 4314.1148
153 Bosnia and Herzegovina 1992 4256013 Europe 72.178 2546.7814
154 Bosnia and Herzegovina 1997 3607000 Europe 73.244 4766.3559
155 Bosnia and Herzegovina 2002 4165416 Europe 74.090 6018.9752
156 Bosnia and Herzegovina 2007 4552198 Europe 74.852 7446.2988
157 Botswana 1952 442308 Africa 47.622 851.2411
158 Botswana 1957 474639 Africa 49.618 918.2325
159 Botswana 1962 512764 Africa 51.520 983.6540
160 Botswana 1967 553541 Africa 53.298 1214.7093
161 Botswana 1972 619351 Africa 56.024 2263.6111
162 Botswana 1977 781472 Africa 59.319 3214.8578
163 Botswana 1982 970347 Africa 61.484 4551.1421
164 Botswana 1987 1151184 Africa 63.622 6205.8839
165 Botswana 1992 1342614 Africa 62.745 7954.1116
166 Botswana 1997 1536536 Africa 52.556 8647.1423
[ reached 'max' / getOption("max.print") -- omitted 1538 rows ]While you get to see what is in the object, perhaps it’s giving us a
little too much information… So let’s permanently coerce
gapminder as a tibble for the rest of the lessons.
gapminder = as_tibble(gapminder)Glimpse
Similar to str(), which is in base R,
glimpse() is a dplyr function that (as the
name suggests) gives a glimpse of the data frame.
glimpse(gapminder)Rows: 1,704
Columns: 6
$ country <chr> "Afghanistan", "Afghanistan", "Afghanistan", "Afghanistan", "Afghanistan", "Afghanistan", "Afghanistan", "Afghanistan", "Afghanistan", "Afghanistan", "Afghanist…
$ year <int> 1952, 1957, 1962, 1967, 1972, 1977, 1982, 1987, 1992, 1997, 2002, 2007, 1952, 1957, 1962, 1967, 1972, 1977, 1982, 1987, 1992, 1997, 2002, 2007, 1952, 1957, 1962…
$ pop <dbl> 8425333, 9240934, 10267083, 11537966, 13079460, 14880372, 12881816, 13867957, 16317921, 22227415, 25268405, 31889923, 1282697, 1476505, 1728137, 1984060, 226355…
$ continent <chr> "Asia", "Asia", "Asia", "Asia", "Asia", "Asia", "Asia", "Asia", "Asia", "Asia", "Asia", "Asia", "Europe", "Europe", "Europe", "Europe", "Europe", "Europe", "Eur…
$ lifeExp <dbl> 28.801, 30.332, 31.997, 34.020, 36.088, 38.438, 39.854, 40.822, 41.674, 41.763, 42.129, 43.828, 55.230, 59.280, 64.820, 66.220, 67.690, 68.930, 70.420, 72.000, …
$ gdpPercap <dbl> 779.4453, 820.8530, 853.1007, 836.1971, 739.9811, 786.1134, 978.0114, 852.3959, 649.3414, 635.3414, 726.7341, 974.5803, 1601.0561, 1942.2842, 2312.8890, 2760.19…In the above output, we can already gather some information about
gapminder, such as the number of rows and columns, column
names, type of vector in the columns, and the first few entries of each
column.
Select
To select columns of a data frame, use select(). The
first argument to this function is the data frame
(gapminder), and the subsequent arguments are the columns
to keep.
select(gapminder, country, year, gdpPercap)# A tibble: 1,704 × 3
country year gdpPercap
<chr> <int> <dbl>
1 Afghanistan 1952 779.
2 Afghanistan 1957 821.
3 Afghanistan 1962 853.
4 Afghanistan 1967 836.
5 Afghanistan 1972 740.
6 Afghanistan 1977 786.
7 Afghanistan 1982 978.
8 Afghanistan 1987 852.
9 Afghanistan 1992 649.
10 Afghanistan 1997 635.
# … with 1,694 more rowsTo select all columns except certain ones, put a “-” in front of the variable to exclude it.
select(gapminder, -lifeExp)# A tibble: 1,704 × 5
country year pop continent gdpPercap
<chr> <int> <dbl> <chr> <dbl>
1 Afghanistan 1952 8425333 Asia 779.
2 Afghanistan 1957 9240934 Asia 821.
3 Afghanistan 1962 10267083 Asia 853.
4 Afghanistan 1967 11537966 Asia 836.
5 Afghanistan 1972 13079460 Asia 740.
6 Afghanistan 1977 14880372 Asia 786.
7 Afghanistan 1982 12881816 Asia 978.
8 Afghanistan 1987 13867957 Asia 852.
9 Afghanistan 1992 16317921 Asia 649.
10 Afghanistan 1997 22227415 Asia 635.
# … with 1,694 more rowsdplyr also provides useful functions to select columns
based on their names. For instance, ends_with() allows you
to select columns that ends with specific letters. For instance, if you
wanted to select columns that end with the letter “p”:
select(gapminder, ends_with("p"))# A tibble: 1,704 × 3
pop lifeExp gdpPercap
<dbl> <dbl> <dbl>
1 8425333 28.8 779.
2 9240934 30.3 821.
3 10267083 32.0 853.
4 11537966 34.0 836.
5 13079460 36.1 740.
6 14880372 38.4 786.
7 12881816 39.9 978.
8 13867957 40.8 852.
9 16317921 41.7 649.
10 22227415 41.8 635.
# … with 1,694 more rowsChallenge
Create a table that contains all the columns with the letter “e” and column “country”, without columns “lifeExp”. Hint: look at the help function
tidyselect::ends_with()we’ve just covered.
Solution
select(gapminder, contains("e"), -lifeExp, country)# A tibble: 1,704 × 4
year continent gdpPercap country
<int> <chr> <dbl> <chr>
1 1952 Asia 779. Afghanistan
2 1957 Asia 821. Afghanistan
3 1962 Asia 853. Afghanistan
4 1967 Asia 836. Afghanistan
5 1972 Asia 740. Afghanistan
6 1977 Asia 786. Afghanistan
7 1982 Asia 978. Afghanistan
8 1987 Asia 852. Afghanistan
9 1992 Asia 649. Afghanistan
10 1997 Asia 635. Afghanistan
# … with 1,694 more rowsFilter
To choose rows based on a logical condition, use
filter():
filter(gapminder, country == 'Nigeria')# A tibble: 12 × 6
country year pop continent lifeExp gdpPercap
<chr> <int> <dbl> <chr> <dbl> <dbl>
1 Nigeria 1952 33119096 Africa 36.3 1077.
2 Nigeria 1957 37173340 Africa 37.8 1101.
3 Nigeria 1962 41871351 Africa 39.4 1151.
4 Nigeria 1967 47287752 Africa 41.0 1015.
5 Nigeria 1972 53740085 Africa 42.8 1698.
6 Nigeria 1977 62209173 Africa 44.5 1982.
7 Nigeria 1982 73039376 Africa 45.8 1577.
8 Nigeria 1987 81551520 Africa 46.9 1385.
9 Nigeria 1992 93364244 Africa 47.5 1620.
10 Nigeria 1997 106207839 Africa 47.5 1625.
11 Nigeria 2002 119901274 Africa 46.6 1615.
12 Nigeria 2007 135031164 Africa 46.9 2014.filter() will keep all the rows that match the
conditions that are provided. Here are a few examples:
# rows for which the country column contains Vietnam or Indonesia
filter(gapminder, country %in% c('Vietnam', 'Indonesia'))# A tibble: 24 × 6
country year pop continent lifeExp gdpPercap
<chr> <int> <dbl> <chr> <dbl> <dbl>
1 Indonesia 1952 82052000 Asia 37.5 750.
2 Indonesia 1957 90124000 Asia 39.9 859.
3 Indonesia 1962 99028000 Asia 42.5 849.
4 Indonesia 1967 109343000 Asia 46.0 762.
5 Indonesia 1972 121282000 Asia 49.2 1111.
6 Indonesia 1977 136725000 Asia 52.7 1383.
7 Indonesia 1982 153343000 Asia 56.2 1517.
8 Indonesia 1987 169276000 Asia 60.1 1748.
9 Indonesia 1992 184816000 Asia 62.7 2383.
10 Indonesia 1997 199278000 Asia 66.0 3119.
# … with 14 more rows# rows with lifeExp greater than or equal to 70
filter(gapminder, lifeExp >= 70)# A tibble: 494 × 6
country year pop continent lifeExp gdpPercap
<chr> <int> <dbl> <chr> <dbl> <dbl>
1 Albania 1982 2780097 Europe 70.4 3631.
2 Albania 1987 3075321 Europe 72 3739.
3 Albania 1992 3326498 Europe 71.6 2497.
4 Albania 1997 3428038 Europe 73.0 3193.
5 Albania 2002 3508512 Europe 75.7 4604.
6 Albania 2007 3600523 Europe 76.4 5937.
7 Algeria 2002 31287142 Africa 71.0 5288.
8 Algeria 2007 33333216 Africa 72.3 6223.
9 Argentina 1987 31620918 Americas 70.8 9140.
10 Argentina 1992 33958947 Americas 71.9 9308.
# … with 484 more rowsfilter() allows you to combine multiple conditions using
the & (AND) logical operator and the |
(OR) logical operator. Such conditions can be arbitrarily complex:
# Get only the data for Germany on or after 1980
filter(gapminder, country == "Germany" & year >= 1980)# A tibble: 6 × 6
country year pop continent lifeExp gdpPercap
<chr> <int> <dbl> <chr> <dbl> <dbl>
1 Germany 1982 78335266 Europe 73.8 22032.
2 Germany 1987 77718298 Europe 74.8 24639.
3 Germany 1992 80597764 Europe 76.1 26505.
4 Germany 1997 82011073 Europe 77.3 27789.
5 Germany 2002 82350671 Europe 78.7 30036.
6 Germany 2007 82400996 Europe 79.4 32170.# Get the data for Australia or Mauritius on or after 1990
filter(gapminder, year >= 1990 & (country == "Australia" | country == 'Mauritius'))# A tibble: 8 × 6
country year pop continent lifeExp gdpPercap
<chr> <int> <dbl> <chr> <dbl> <dbl>
1 Australia 1992 17481977 Oceania 77.6 23425.
2 Australia 1997 18565243 Oceania 78.8 26998.
3 Australia 2002 19546792 Oceania 80.4 30688.
4 Australia 2007 20434176 Oceania 81.2 34435.
5 Mauritius 1992 1096202 Africa 69.7 6058.
6 Mauritius 1997 1149818 Africa 70.7 7426.
7 Mauritius 2002 1200206 Africa 72.0 9022.
8 Mauritius 2007 1250882 Africa 72.8 10957.Challenge
Select all data for countries in Europe between the years 1990 and 2000
Solution
filter(gapminder, continent == 'Europe' & (year >= 1990 & year <= 2000))# A tibble: 60 × 6
country year pop continent lifeExp gdpPercap
<chr> <int> <dbl> <chr> <dbl> <dbl>
1 Albania 1992 3326498 Europe 71.6 2497.
2 Albania 1997 3428038 Europe 73.0 3193.
3 Austria 1992 7914969 Europe 76.0 27042.
4 Austria 1997 8069876 Europe 77.5 29096.
5 Belgium 1992 10045622 Europe 76.5 25576.
6 Belgium 1997 10199787 Europe 77.5 27561.
7 Bosnia and Herzegovina 1992 4256013 Europe 72.2 2547.
8 Bosnia and Herzegovina 1997 3607000 Europe 73.2 4766.
9 Bulgaria 1992 8658506 Europe 71.2 6303.
10 Bulgaria 1997 8066057 Europe 70.3 5970.
# … with 50 more rowsPipes
But what if you wanted to select and filter? We can do this with
pipes (which we saw in the bash lessons). Pipes, are a
fairly recent addition to R. Pipes let you take the output of one
function and send it directly to the next, which is useful when you need
to do many things to the same data set sequentially. Pipes in R look
like %>% (recall they looked like | in
bash) and are available via the magrittr
package, which is installed as part of dplyr. If you use
RStudio, you can type the pipe with Ctrl + Shift +
M if you’re using a PC, or Cmd + Shift
+ M if you’re using a Mac.
# Filter for data from Spain and select year, pop, and lifeExp columns
gapminder %>%
filter(country == "Spain") %>%
select(year, pop, lifeExp)# A tibble: 12 × 3
year pop lifeExp
<int> <dbl> <dbl>
1 1952 28549870 64.9
2 1957 29841614 66.7
3 1962 31158061 69.7
4 1967 32850275 71.4
5 1972 34513161 73.1
6 1977 36439000 74.4
7 1982 37983310 76.3
8 1987 38880702 76.9
9 1992 39549438 77.6
10 1997 39855442 78.8
11 2002 40152517 79.8
12 2007 40448191 80.9In the above code, we use the pipe to send the gapminder
dataset first through filter(), to keep rows where
country matches a particular country, and then through
select() to keep only the year,
pop, and lifeExp columns. Since
%>% takes the object on its left and passes it as the
first argument to the function on its right, we don’t need to explicitly
include the data frame as an argument to the filter() and
select() functions any more.
Some may find it helpful to read the pipe like the word “then”. For
instance, in the above example, we took the data frame
gapminder, then we filtered for rows
where country was Spain, then we
selected the year, pop, and
lifeExp columns, then we showed only the first six
rows. The dplyr functions by themselves
are somewhat simple, but by combining them into linear workflows with
the pipe, we can accomplish more complex manipulations of data
frames.
If we want to create a new object with this smaller version of the data we can do so by assigning it a new name:
spain_gapminder <- gapminder %>%
filter(country == "Spain") %>%
select(year, pop, lifeExp)This new object includes all of the data from this sample. Let’s look at just the first six rows to confirm it’s what we want:
spain_gapminder# A tibble: 12 × 3
year pop lifeExp
<int> <dbl> <dbl>
1 1952 28549870 64.9
2 1957 29841614 66.7
3 1962 31158061 69.7
4 1967 32850275 71.4
5 1972 34513161 73.1
6 1977 36439000 74.4
7 1982 37983310 76.3
8 1987 38880702 76.9
9 1992 39549438 77.6
10 1997 39855442 78.8
11 2002 40152517 79.8
12 2007 40448191 80.9Similar to head() and tail() functions, we
can also look at the first or last six rows using tidyverse function
slice(). Slice is a more versatile function that allows
users to specify a range to view:
spain_gapminder %>% slice(1:6)# A tibble: 6 × 3
year pop lifeExp
<int> <dbl> <dbl>
1 1952 28549870 64.9
2 1957 29841614 66.7
3 1962 31158061 69.7
4 1967 32850275 71.4
5 1972 34513161 73.1
6 1977 36439000 74.4spain_gapminder %>% slice(7:11)# A tibble: 5 × 3
year pop lifeExp
<int> <dbl> <dbl>
1 1982 37983310 76.3
2 1987 38880702 76.9
3 1992 39549438 77.6
4 1997 39855442 78.8
5 2002 40152517 79.8Exercise: Pipe and filter
Starting with the
gapminderdata frame, use pipes to subset the data to include only observations from Panama, where the year is at least 1980. Showing only the 4th through 6th rows of columnscountry,year, andgdpPercap.
Solution
gapminder %>%
filter(country == "Panama" & year >= 1980) %>%
slice(4:6) %>%
select(country, year, gdpPercap)# A tibble: 3 × 3
country year gdpPercap
<chr> <int> <dbl>
1 Panama 1997 7114.
2 Panama 2002 7356.
3 Panama 2007 9809.Mutate
Frequently you’ll want to create new columns based on the values in
existing columns, for example to do unit conversions or find the ratio
of values in two columns. For this we’ll use the dplyr
function mutate().
We have a column titled “gdpPercap” and “pop”. We could use these two columns to compute the “total_gdp” for each country/year observation. By multiplying the entries per-row.
Let’s add a column (total_gdp) to our
gapminder data frame that shows the total GDP for the
country in the corresponding year.
gapminder %>% mutate(total_gdp = gdpPercap * pop)# A tibble: 1,704 × 7
country year pop continent lifeExp gdpPercap total_gdp
<chr> <int> <dbl> <chr> <dbl> <dbl> <dbl>
1 Afghanistan 1952 8425333 Asia 28.8 779. 6567086330.
2 Afghanistan 1957 9240934 Asia 30.3 821. 7585448670.
3 Afghanistan 1962 10267083 Asia 32.0 853. 8758855797.
4 Afghanistan 1967 11537966 Asia 34.0 836. 9648014150.
5 Afghanistan 1972 13079460 Asia 36.1 740. 9678553274.
6 Afghanistan 1977 14880372 Asia 38.4 786. 11697659231.
7 Afghanistan 1982 12881816 Asia 39.9 978. 12598563401.
8 Afghanistan 1987 13867957 Asia 40.8 852. 11820990309.
9 Afghanistan 1992 16317921 Asia 41.7 649. 10595901589.
10 Afghanistan 1997 22227415 Asia 41.8 635. 14121995875.
# … with 1,694 more rowsExercise
There is data for a lot of countries and years, so let’s look just at the results of the United States by adding the correct line to the above code.
Solution
gapminder %>%
mutate(total_gdp = gdpPercap * pop) %>%
filter(country == 'United States')# A tibble: 12 × 7
country year pop continent lifeExp gdpPercap total_gdp
<chr> <int> <dbl> <chr> <dbl> <dbl> <dbl>
1 United States 1952 157553000 Americas 68.4 13990. 2.20e12
2 United States 1957 171984000 Americas 69.5 14847. 2.55e12
3 United States 1962 186538000 Americas 70.2 16173. 3.02e12
4 United States 1967 198712000 Americas 70.8 19530. 3.88e12
5 United States 1972 209896000 Americas 71.3 21806. 4.58e12
6 United States 1977 220239000 Americas 73.4 24073. 5.30e12
7 United States 1982 232187835 Americas 74.6 25010. 5.81e12
8 United States 1987 242803533 Americas 75.0 29884. 7.26e12
9 United States 1992 256894189 Americas 76.1 32004. 8.22e12
10 United States 1997 272911760 Americas 76.8 35767. 9.76e12
11 United States 2002 287675526 Americas 77.3 39097. 1.12e13
12 United States 2007 301139947 Americas 78.2 42952. 1.29e13Group by and summarize
Many data analysis tasks can be approached using the
“split-apply-combine” paradigm: split the data into groups, apply some
analysis to each group, and then combine the results. dplyr
makes this very easy through the use of the group_by()
function, which splits the data into groups. When the data is grouped in
this way summarize() can be used to collapse each group
into a single-row summary. summarize() does this by
applying an aggregating or summary function to each group. For example,
if we wanted to group by continent and find the number of rows of data
for each continent, we would do:
gapminder %>%
group_by(continent) %>%
summarize(n())# A tibble: 5 × 2
continent `n()`
<chr> <int>
1 Africa 624
2 Americas 300
3 Asia 396
4 Europe 360
5 Oceania 24Notice this is the same results as when we ran
summary(gapminder$continent).
It can be a bit tricky at first, but we can imagine splitting the data frame by groups and applying a certain function to summarize the data.
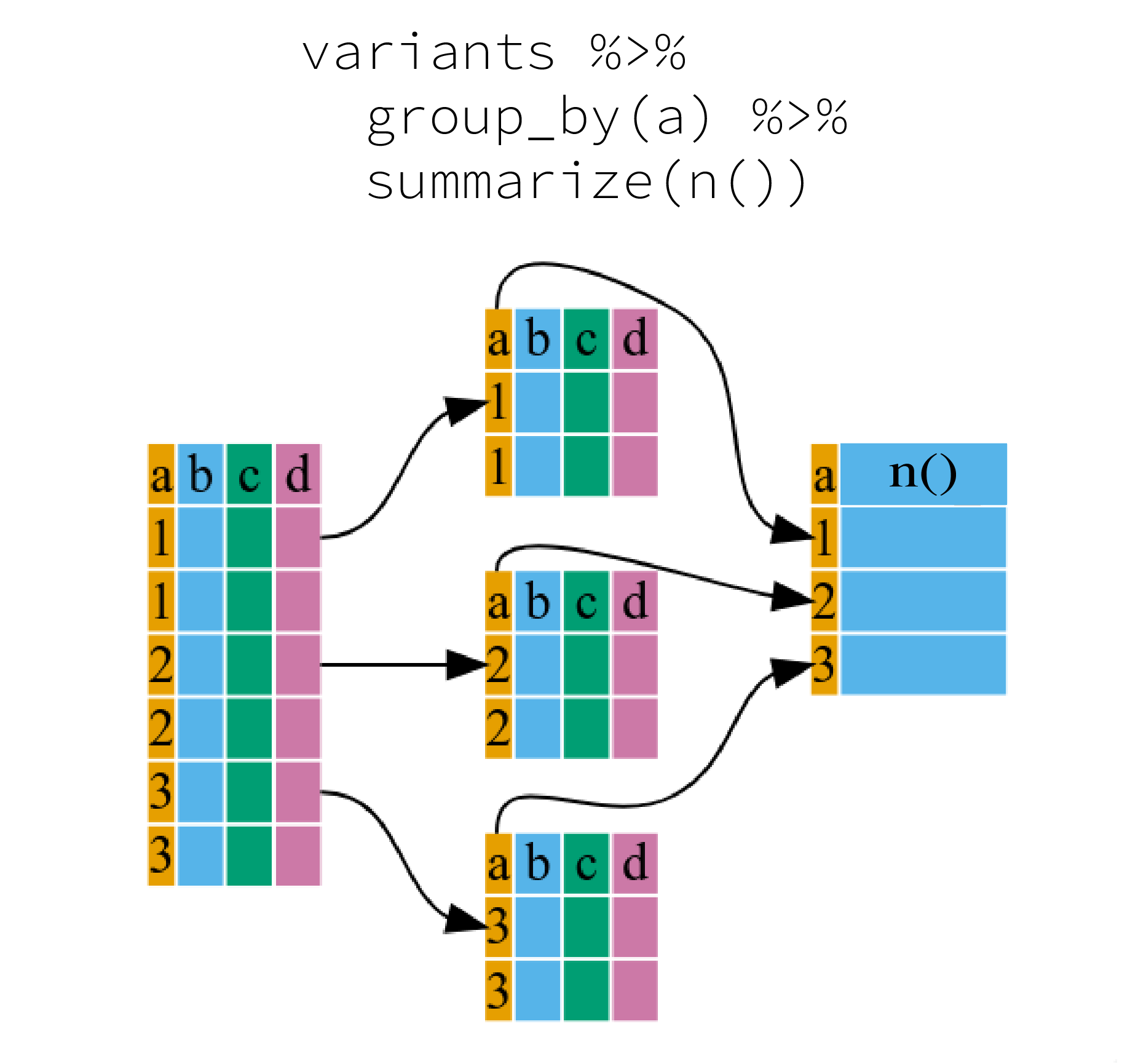
Here the summary function used was n() to find the count
for each group. Since this is a quite a common operation, there is a
simpler method called tally():
gapminder %>%
group_by(country) %>%
tally()# A tibble: 142 × 2
country n
<chr> <int>
1 Afghanistan 12
2 Albania 12
3 Algeria 12
4 Angola 12
5 Argentina 12
6 Australia 12
7 Austria 12
8 Bahrain 12
9 Bangladesh 12
10 Belgium 12
# … with 132 more rowsTo show that there are many ways to achieve the same results, there
is another way to approach this, which bypasses group_by()
using the function count():
gapminder %>% count(country)# A tibble: 142 × 2
country n
<chr> <int>
1 Afghanistan 12
2 Albania 12
3 Algeria 12
4 Angola 12
5 Argentina 12
6 Australia 12
7 Austria 12
8 Bahrain 12
9 Bangladesh 12
10 Belgium 12
# … with 132 more rowsWe can also apply many other functions to individual columns to get
other summary statistics. For example,we can use built-in functions like
mean(), median(), min(), and
max(). These are called “built-in functions” because they
come with R and don’t require that you install any additional packages.
By default, all R functions operating on vectors that contains
missing data will return NA. It’s a way to make sure that users
know they have missing data, and make a conscious decision on how to
deal with it. When dealing with simple statistics like the mean, the
easiest way to ignore NA (the missing data) is to use
na.rm = TRUE (rm stands for remove).
So to view the mean, median, maximum, and minimum
gdpPercap for each country:
gapminder %>%
group_by(country) %>%
summarize(
min_gpc = min(gdpPercap),
mean_gpc = mean(gdpPercap),
median_gpc = median(gdpPercap),
max_gpc = max(gdpPercap))# A tibble: 142 × 5
country min_gpc mean_gpc median_gpc max_gpc
<chr> <dbl> <dbl> <dbl> <dbl>
1 Afghanistan 635. 803. 803. 978.
2 Albania 1601. 3255. 3253. 5937.
3 Algeria 2449. 4426. 4854. 6223.
4 Angola 2277. 3607. 3265. 5523.
5 Argentina 5911. 8956. 9069. 12779.
6 Australia 10040. 19981. 18906. 34435.
7 Austria 6137. 20412. 20673. 36126.
8 Bahrain 9867. 18078. 18780. 29796.
9 Bangladesh 630. 818. 704. 1391.
10 Belgium 8343. 19901. 20049. 33693.
# … with 132 more rowsThe pivot functions
It can be useful to transform the “long” tidy format, into the wide
format. This transformation can be done with the
pivot_wider() function provided by the tidyr
package (also part of the tidyverse).
pivot_wider() takes a data frame as the first argument,
and two arguments: the column name that will become the columns and the
column name that will become the cells in the wide data. Let’s create a
wide format table with rows for each country, columns for each year, and
data values being the lifeExp.
gapminder_wide <- gapminder %>%
select(country, lifeExp, year) %>%
group_by(country) %>%
pivot_wider(names_from = year, values_from = lifeExp)
gapminder_wide# A tibble: 142 × 13
# Groups: country [142]
country `1952` `1957` `1962` `1967` `1972` `1977` `1982` `1987` `1992` `1997` `2002` `2007`
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Afghanistan 28.8 30.3 32.0 34.0 36.1 38.4 39.9 40.8 41.7 41.8 42.1 43.8
2 Albania 55.2 59.3 64.8 66.2 67.7 68.9 70.4 72 71.6 73.0 75.7 76.4
3 Algeria 43.1 45.7 48.3 51.4 54.5 58.0 61.4 65.8 67.7 69.2 71.0 72.3
4 Angola 30.0 32.0 34 36.0 37.9 39.5 39.9 39.9 40.6 41.0 41.0 42.7
5 Argentina 62.5 64.4 65.1 65.6 67.1 68.5 69.9 70.8 71.9 73.3 74.3 75.3
6 Australia 69.1 70.3 70.9 71.1 71.9 73.5 74.7 76.3 77.6 78.8 80.4 81.2
7 Austria 66.8 67.5 69.5 70.1 70.6 72.2 73.2 74.9 76.0 77.5 79.0 79.8
8 Bahrain 50.9 53.8 56.9 59.9 63.3 65.6 69.1 70.8 72.6 73.9 74.8 75.6
9 Bangladesh 37.5 39.3 41.2 43.5 45.3 46.9 50.0 52.8 56.0 59.4 62.0 64.1
10 Belgium 68 69.2 70.2 70.9 71.4 72.8 73.9 75.4 76.5 77.5 78.3 79.4
# … with 132 more rowsThe opposite operation of pivot_wider() is taken care by
pivot_longer(). We specify the names of the new columns,
and here add -country as this column shouldn’t be affected
by the reshaping:
gapminder_wide %>%
pivot_longer(-country, names_to = "year", values_to = "lifeExp")# A tibble: 1,704 × 3
# Groups: country [142]
country year lifeExp
<chr> <chr> <dbl>
1 Afghanistan 1952 28.8
2 Afghanistan 1957 30.3
3 Afghanistan 1962 32.0
4 Afghanistan 1967 34.0
5 Afghanistan 1972 36.1
6 Afghanistan 1977 38.4
7 Afghanistan 1982 39.9
8 Afghanistan 1987 40.8
9 Afghanistan 1992 41.7
10 Afghanistan 1997 41.8
# … with 1,694 more rowsTip: Why pivot functions?
Imagine that you want to put the gapminder data into a paper where the rows are countries, the columns are years, and the data values are life expectancy. In it’s currrent form, that data is long, and is not a natural format for a paper. The
pivot_wider()function will transform the data into a more compact form suitable for a paper.Now, imagine you want to download and use data from a table in a paper. Say the rows are countries, the columns are years, and the data values are life expectancy. But you want to add some additional data based on the country and year that you have collected. The wide format of the data doesn’t easily let you do this, but the long format does. In which case, the
pivot_longer()function will help you transform the data so you can amend it for your purposes.
Tip: Trial and error with pivot functions
The pivot functions will often require some trial and error to arrive at the desired output.
Resources
The figure was adapted from the Software Carpentry lesson, R for Reproducible Scientific Analysis↩︎