Intro to Single Cell RNA-Seq Workshop
UM Bioinformatics Core Workshop Team
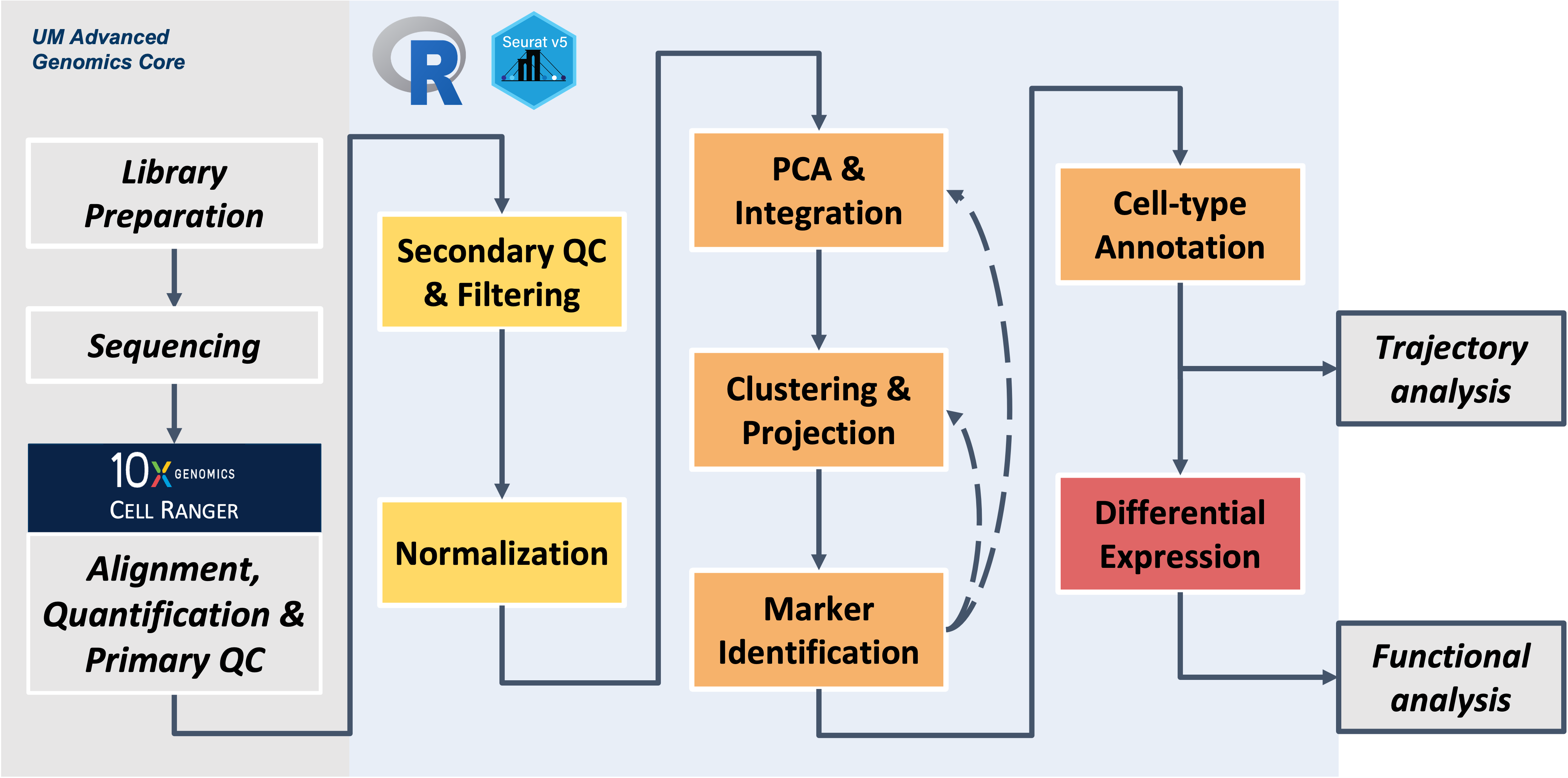
Wrapping up
We hope you now have more familiarity with key concepts, data types, tools, and how they all connect to enable single-cell gene expression analysis from RNA-Seq data.
Housekeeping
Please take our optional post-workshop survey (5-10 minutes).
We will email you a link to the final session recordings next week.
The website/notes for this workshop will be available.
The UM Bioinformatics Core Workshop Slack channel content will be available for 90 days.
Can I continue to use the RStudio environments we used in the workshop?
The RStudio workshop compute environment will be available through 10/24/2025.
- Please save all your R scripts now so that we can “right-size” the compute environment immediately following today’s workshop session.
You can download files from the workshop environment from your terminal/command line window as below. (You will need to substitute your actual workshop username and type workshop password when prompted.)
# download workshop files ------------------------------------------------- mkdir intro_scrnaseq_workshop cd intro_scrnaseq_workshop scp -r YOUR_USERNAME@bfx-workshop01.med.umich.edu:"ISC_R*" .- Note that the full download of the R data is about 8Gb, so depending on your internet speeds it could take a while. (We do not recommend you download the full set of Cell Ranger outputs.)
Where else could I use RStudio?
- See Advanced setup instructions for details on how to
install RStudio and required packages on your own computer. Please note:
- Installing bioinformatics software is often non-trivial.
- For typical data, Cell Ranger steps (reviewed Day 1) assume your computer has powerful compute (many CPUs and lots of RAM) and sizable storage capacity. (i.e. it’s impractical to run these on your laptop.)
- PositCloud allows you to run R-Studio over the web for free. Click here to login to PositCloud and launch an R session. (UM users can login using their umich.edu Google account.) Note: A full single-cell analysis will require you to upgrade beyond the free tier.
- UMich users can run RStudio on the UM Great Lakes HPC.
Could I download Seurat inputs used in the workshop?
The Seurat inputs (cellranger triples and dbcells files) used in this workshop can be installed locally like so:
# download Seurat inputs -------------------------------------------------- mkdir -p intro_scrnaseq_workshop/ISC_R cd intro_scrnaseq_workshop/ISC_R # Use curl to download a ~3 Gb tarball # We'll use evironment variables to avoid extremely long command lines source_url="https://umich-brcf-bioinf-workshop.s3.us-east-1.amazonaws.com" source_file="ISC/workshop_isc_inputs-20251015.tgz" curl -o workshop_isc_inputs.tgz ${source_url}/${source_file} # tar unpacks the tarball into directories tar xzvf workshop_isc_inputs.tgz # Since we have unpacked the tarball, we can remove it rm workshop_isc_inputs.tgz
Can you recommend other relevant workshops or tutorials?
- The University of Michigan Bioinformatics Core regularly hosts workshops.
- Intro lessons and workshops in Bash / Git / R / Python :
- Software Carpentry
- The Carpentries at the University of Michigan host several introductory workshops each year following curricula that are developed collaboratively and using the latest research into best practices for teaching computer science content. All workshops are offered for free to learners.
How can I learn more about computational research at University of Michigan?
- UM CoderSpaces “office hours” and UM CoderSpaces Slack workspace. (See “Useful Resources” section in CoderSpaces page for instructions on how to join and access the CoderSpaces Slack workspace.)
- Advanced Research Computing
- Research Computing Package
- Upcoming UM Advanced Research Computing workshops
- Videos on getting started with Great Lakes (Requires UM login)
Thank you to our sponsors

Thank you to/from the workshop team
 |
 |
 |
 |
|---|---|---|---|
| Chris | Marci | Raymond | Dana |
 |
 |
 |
|
| Weisheng | Ford | Tricia |
Thank you for participating in our workshop. We welcome your questions and feedback now and in the future.
Bioinformatics Workshop Team
bioinformatics-workshops@umich.edu
UM BRCF Bioinformatics Core