Welcome to Software Carpentry
Overview
Teaching: 15 min
Exercises: 0 minQuestions
What is The Carpentries?
What will the workshop cover?
What else do I need to know about the workshop?
Objectives
Introduce The Carpentries.
Go over logistics.
Introduce the workshop goals.
What is The Carpentries / UM Carpentries?
The Carpentries is a global organization whose mission is to teach researchers, and others, the basics of coding so that you can use it in your own work. We believe everyone can learn to code, and that a lot of you will find it very useful for things such as data analysis and plotting.
Our workshops are targeted to absolute beginners, and we expect that you have zero coding experience coming in. That being said, you’re welcome to attend a workshop if you already have a coding background but want to learn more!
The Carpentries at the University of Michigan is local group of data and software enthusiasts that regularly present workshops for UM groups and the community at large. Let us know if you have interest in hosting a workshop for your group, participating as a helper in a future workshop, or becoming trained as an instuctor.
Code of Conduct
To provide an inclusive learning environment, we follow The Carpentries Code of Conduct. We expect that instructors, helpers, and learners abide by this code of conduct, including practicing the following behaviors:
- Use welcoming and inclusive language.
- Be respectful of different viewpoints and experiences.
- Gracefully accept constructive criticism.
- Focus on what is best for the community.
- Show courtesy and respect towards other community members.
You can report any violations to the Code of Conduct by filling out this form.
Introducing the instructors and helpers
Now that you know a little about The Carpentries as an organization, the instructors and helpers will introduce themselves and what they’ll be teaching/helping with.
Using Zoom and Slack
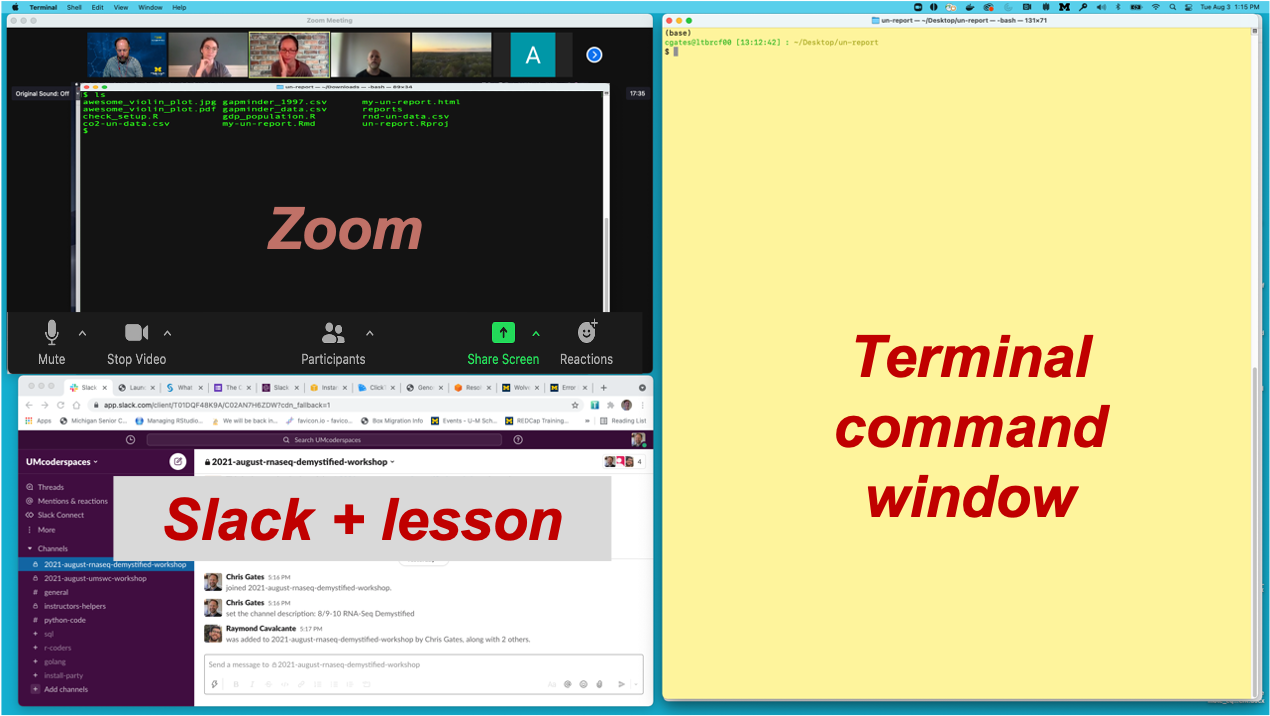
Arranging your screens
It is important that you can see:
- Zoom (instructor’s shared screen + reactions)
- Slack
- R/Studio or terminal/command window
Using Zoom
-
Zoom controls are at the bottom of the Zoom window:

- To minimize distractions, we encourage participants to keep their audio muted (unless actively asking a question).
- To maximize engagement, we encourage participants to keep their video on.
- Slack works better than Zoom’s Chat function so avoid Zoom Chat for now.
- You can enable transcription subtitles for your view.
- We will be using Breakout Rooms occasionally for ad-hoc 1-1 helper support. We will review this in detail together in a few minutes.
- Zoom’s “non-verbal controls” are a useful way to interact
- Depending on your version of Zoom you can access these either
- in the Reactions button on you main Zoom window
- at the bottom of the Participant pane
- Depending on your version of Zoom you can access these either
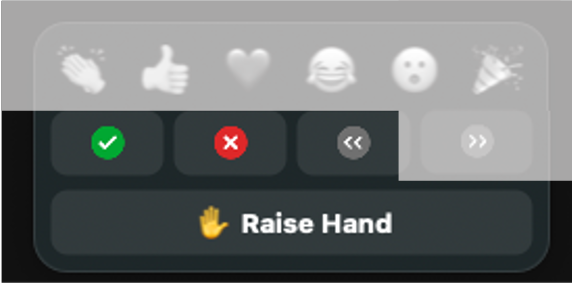
- Raise Hand to request clarification or ask a question. (Same an in-person workshop.)
- Gray chevron [Slow down / Give me a moment] when you need more time to complete an exercise or you would like the instructor to repeat an idea
- Instructors will use Green check and Red X to poll the group
at checkpoints along the way.
Exercise: Use Zoom non-verbals
- Everyone use Zoom to raise your hand.

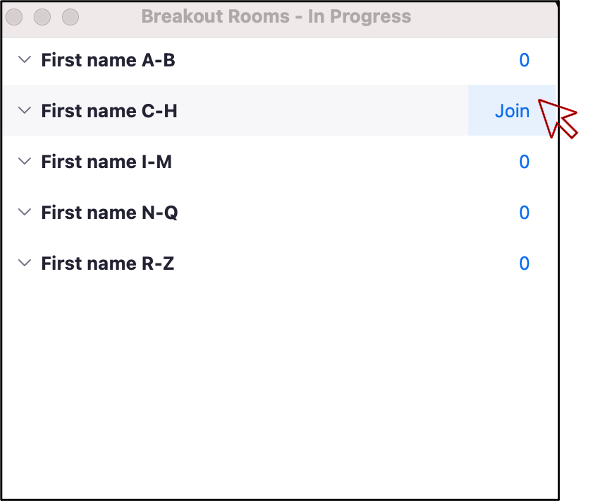
Exercise: Using Zoom Breakout Rooms
Take a moment to briefly introduce yourself (name, dept/lab, area of study) in a breakout room.
- Zoom: Click Breakout Rooms
- Find the room corresponding to the first letter of your first name
- Hover over the number to the right and click Join.
- When you have completed introductions, you can leave the breakout room to rejoin the main room.
Using Slack
- Slack can be used to communicate to the group or to individuals and has a few features/behaviors that we prefer over Zoom’s Chat functionality.
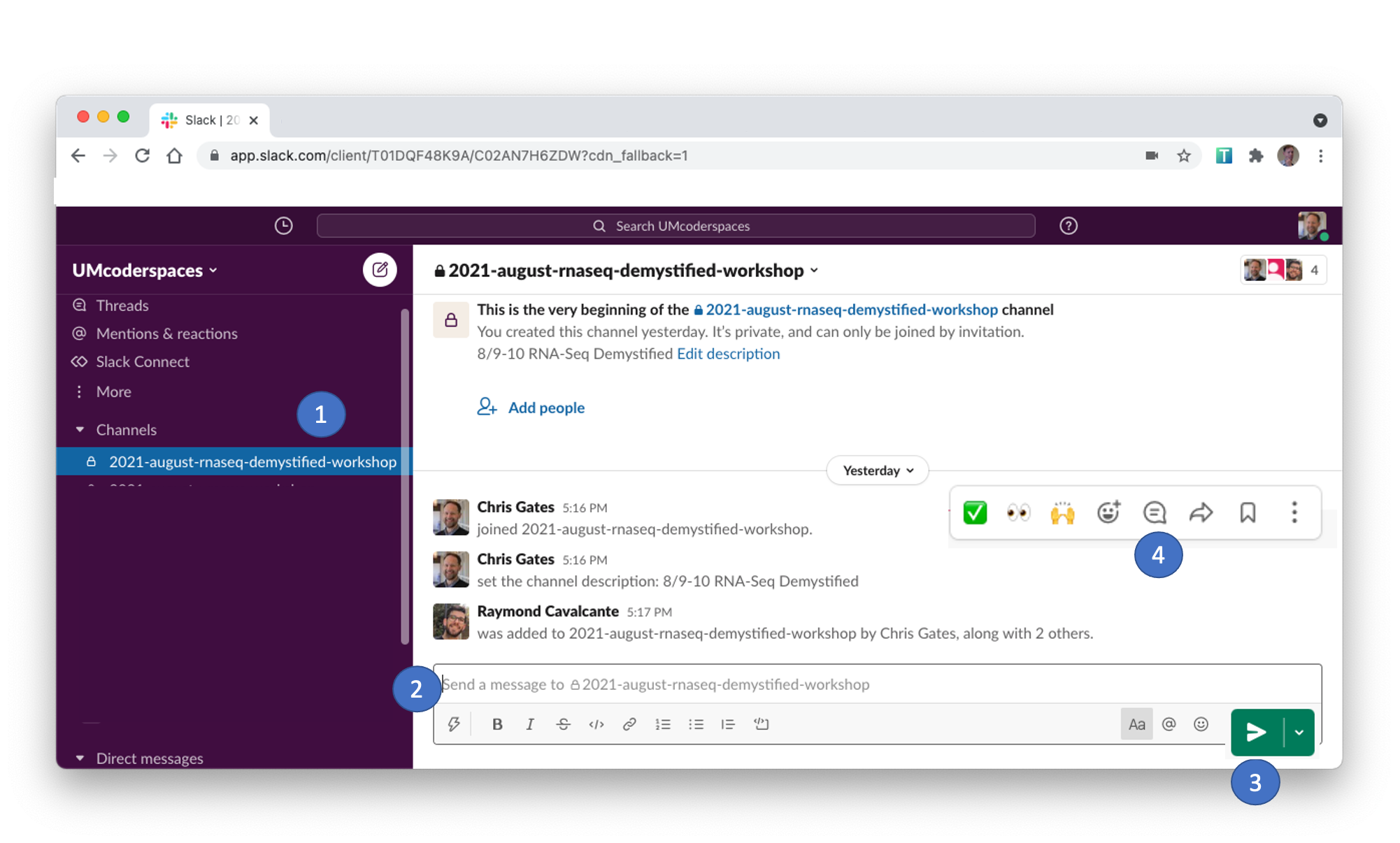
- Slack messages will be posted to the 2021-october-umswc-workshop channel.
Click on the channel in the left pane (1) to select this channel. - You can type in the message field (2); click send (3) to post your message to everyone.
- Helpers will respond in a Slack thread (or pose the question to the instructor)
- You can respond in a message thread by hovering over a message to trigger the message menu and clicking the speech bubble (4).
Exercise: Responding in Slack thread
What is one thing you hope to learn today or tomorrow?
Review of Key communication patterns
| Syntax |  |
 |
|---|---|---|
| “I have an urgent question” |  |
Post a question |
| “I have a general question” |  |
Post a question |
| “I’m stuck / I need a hand” | Post a note | |
| Instructor check-in |  (OK) (OK) or  (Wha?) (Wha?) |
|
| Instructor Slack question | Respond in Slack thread |
Exercise: Group checkpoint
- Using Zoom, give me a green-check if you feel like you understand
communication patterns or red-X if you need clarification.
The “goal” of the workshop
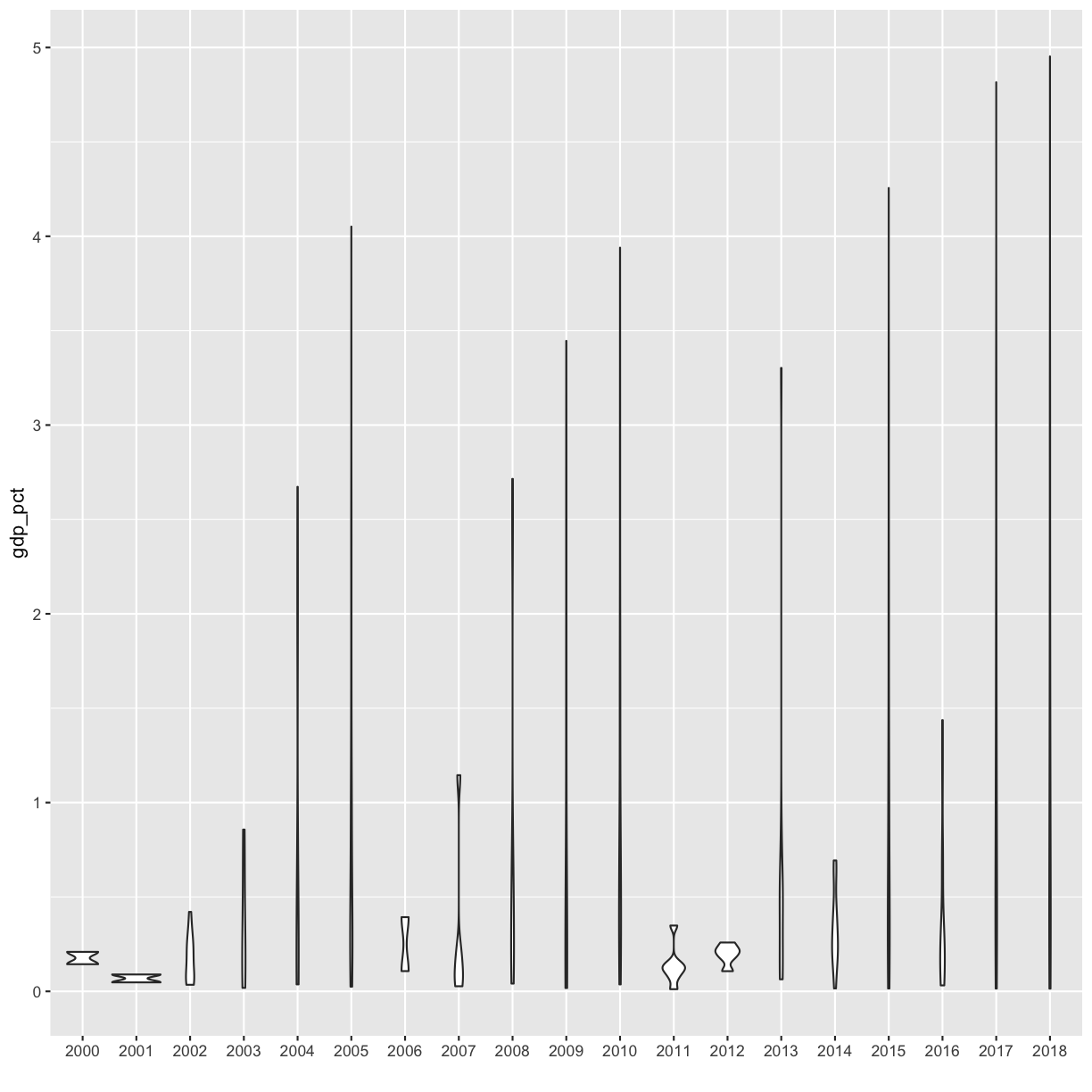
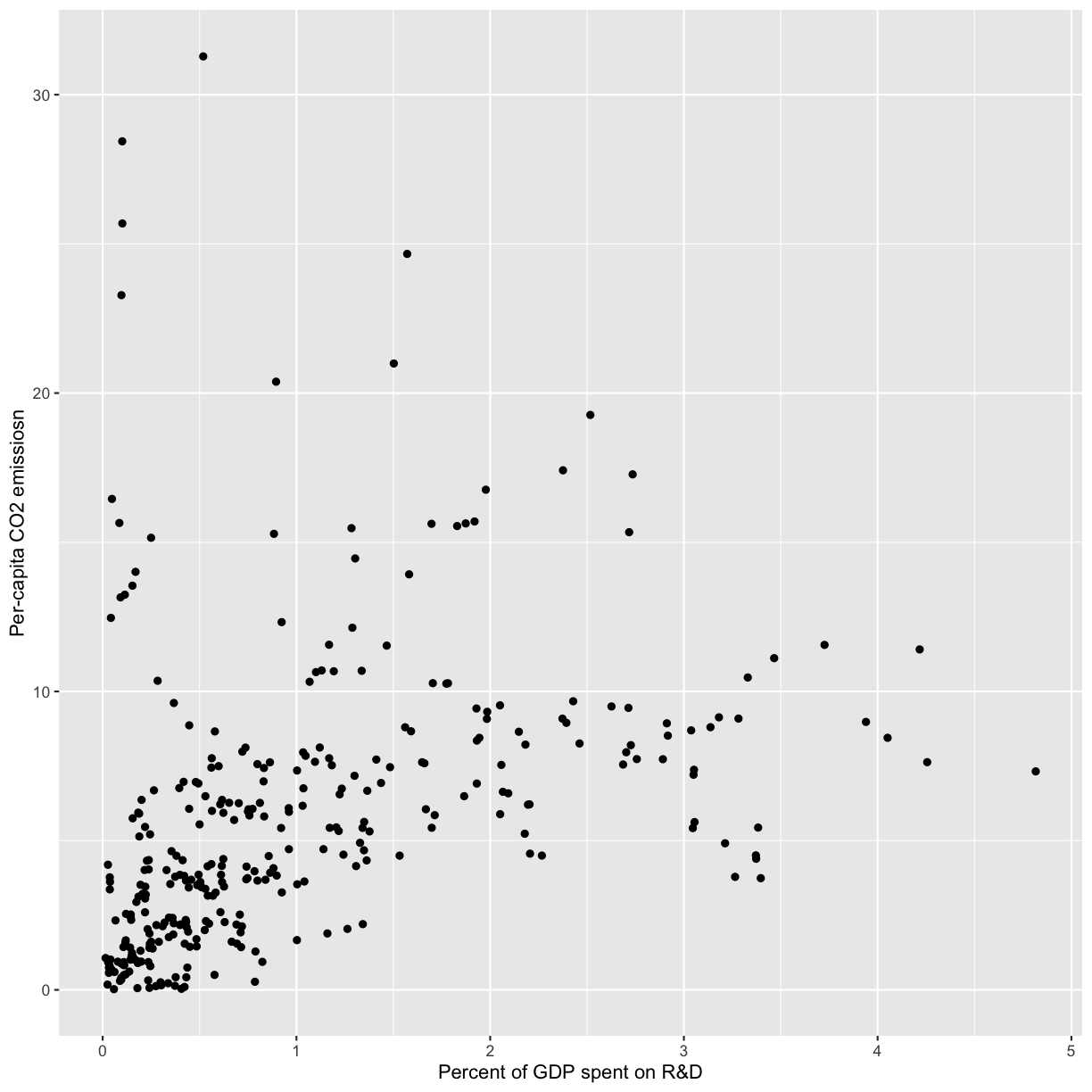
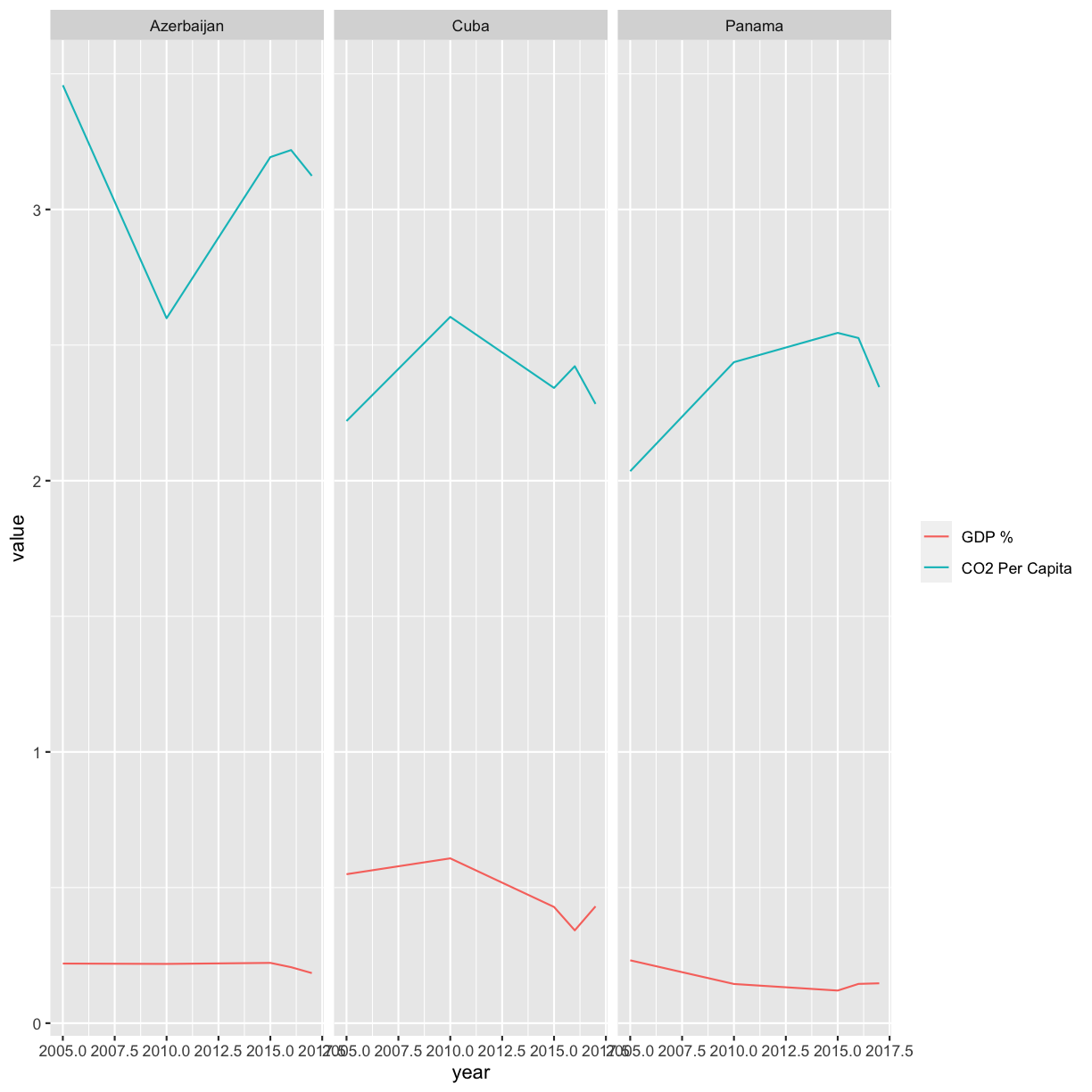
Now that we all know each other, let’s learn a bit more about why we’re here. Our goal is to write a report to the United Nations on the relationship between GDP, life expectancy, and CO2 emissions. In other words, we are going to analyze how countries’ economic strength or weakness may be related to public health status and climate pollution, respectively.
To get to that point, we’ll need to learn how to manage data, make plots, and generate reports. The next section discusses in more detail exactly what we will cover.
What will the workshop cover?
This workshop will introduce you to some of the programs used everyday in computational workflows in diverse fields: microbiology, statistics, neuroscience, genetics, the social and behavioral sciences, such as psychology, economics, public health, and many others.
A workflow is a set of steps to read data, analyze it, and produce numerical and graphical results to support an assertion or hypothesis encapsulated into a set of computer files that can be run from scratch on the same data to obtain the same results. This is highly desirable in situations where the same work is done repeatedly – think of processing data from an annual survey, or results from a high-throughput sequencer on a new sample. It is also desirable for reproducibility, which enables you and other people to look at what you did and produce the same results later on. It is increasingly common for people to publish scientific articles along with the data and computer code that generated the results discussed within them.
The programs to be introduced are:
- R, RStudio, and R Markdown: a statistical analysis and data management program, a graphical interface to it, and a method for writing reproducible reports. We’ll use it to manage data and make pretty plots!
- Git: a program to help you keep track of changes to your programs over time.
- GitHub: a web application that makes sharing your programs and working on them with others much easier. It can also be used to generate a citable reference to your computer code.
- The Unix shell (command line): A tool that is extremely useful for managing both data and program files and chaining together discrete steps in your workflow (automation).
We will not try to make you an expert or even proficient with any of them, but we hope to demonstrate the basics of controlling your code, automating your work, and creating reproducible programs. We also hope to provide you with some fundamentals that you can incorporate in your own work.
At the end, we provide links to resources you can use to learn about these topics in more depth than this workshop can provide.
Other miscellaneous things
If you’re in person, we’ll tell you where the bathrooms are! If you’re virtual we hope you know. :) Let us know if there are any accommodations we can provide to help make your learning experience better!
Key Points
We follow The Carpentries Code of Conduct.
Our goal is to generate a shareable and reproducible report by the end of the workshop.
This lesson content is targeted to absolute beginners with no coding experience.
R for Plotting
Overview
Teaching: 120 min
Exercises: 30 minQuestions
What are R and R Studio?
How do I read data into R?
What are geometries and aesthetics?
How can I use R to create and save professional data visualizations?
Objectives
To become oriented with R and R Studio.
To be able to read in data from csv files.
To create plots with both discrete and continuous variables.
To understand mapping and layering using
ggplot2.To be able to modify a plot’s color, theme, and axis labels.
To be able to save plots to a local directory.
Contents
- Introduction to R and RStudio
- Loading and reviewing data
- Understanding commands
- Creating our first plot
- Plotting for data exploration
- [Bonus topics!]
- Glossary of terms
Bonus: why learn to program?
Share why you’re interested in learning how to code.
Solution:
There are lots of different reasons, including to perform data analysis and generate figures. I’m sure you have morespecific reasons for why you’d like to learn!
Introduction to R and RStudio
In this session we will be testing the hypothesis that a country’s life expectancy is related to the total value of its finished goods and services, also known as the Gross Domestic Product (GDP). To test this hypothesis, we’ll need two things: data and a platform to analyze the data.
You already downloaded the data. But what platform will we use to analyze the data? We have many options!
We could try to use a spreadsheet program like Microsoft Excel or Google sheets that have limited access, less flexibility, and don’t easily allow for things that are critical to “reproducible” research, like easily sharing the steps used to explore and make changes to the original data.
Instead, we’ll use a programming language to test our hypothesis. Today we will use R, but we could have also used Python for the same reasons we chose R (and we teach workshops for both languages). Both R and Python are freely available, the instructions you use to do the analysis are easily shared, and by using reproducible practices, it’s straightforward to add more data or to change settings like colors or the size of a plotting symbol.
But why R and not Python?
There’s no great reason. Although there are subtle differences between the languages, it’s ultimately a matter of personal preference. Both are powerful and popular languages that have very well developed and welcoming communities of scientists that use them. As you learn more about R, you may find things that are annoying in R that aren’t so annoying in Python; the same could be said of learning Python. If the community you work in uses R, then you’re in the right place.
To run R, all you really need is the R program, which is available for computers running the Windows, Mac OS X, or Linux operating systems. You downloaded R while getting set up for this workshop.
To make your life in R easier, there is a great (and free!) program called RStudio that you also downloaded and used during set up. As we work today, we’ll use features that are available in RStudio for writing and running code, managing projects, installing packages, getting help, and much more. It is important to remember that R and RStudio are different, but complementary programs. You need R to use RStudio.
Bonus Exercise: Can you think of a reason you might not want to use RStudio?
Solution:
On some high-performance computer systems (e.g. Amazon Web Services) you typically can’t get a display like RStudio to open. If you’re at the University of Michigan and have access to Great Lakes, then you might want to learn more about resources to run RStudio on Great Lakes.
Tidyverse vs Base R
If you’ve used R before, you may have learned commands that are different than the ones we will be using during this workshop. We will be focusing on functions from the Tidyverse. The “tidyverse” is a collection of R packages that have been designed to work well together and offer many convenient features that do not come with a fresh install of R (aka “base R”). These packages are very popular and have a lot of developer support including many staff members from RStudio. These functions generally help you to write code that is easier to read and maintain. We believe learning these tools will help you become more productive more quickly.
To get started, we’ll spend a little time getting familiar with the RStudio environment and setting it up to suit your tastes. When you start RStudio, you’ll have three panels.
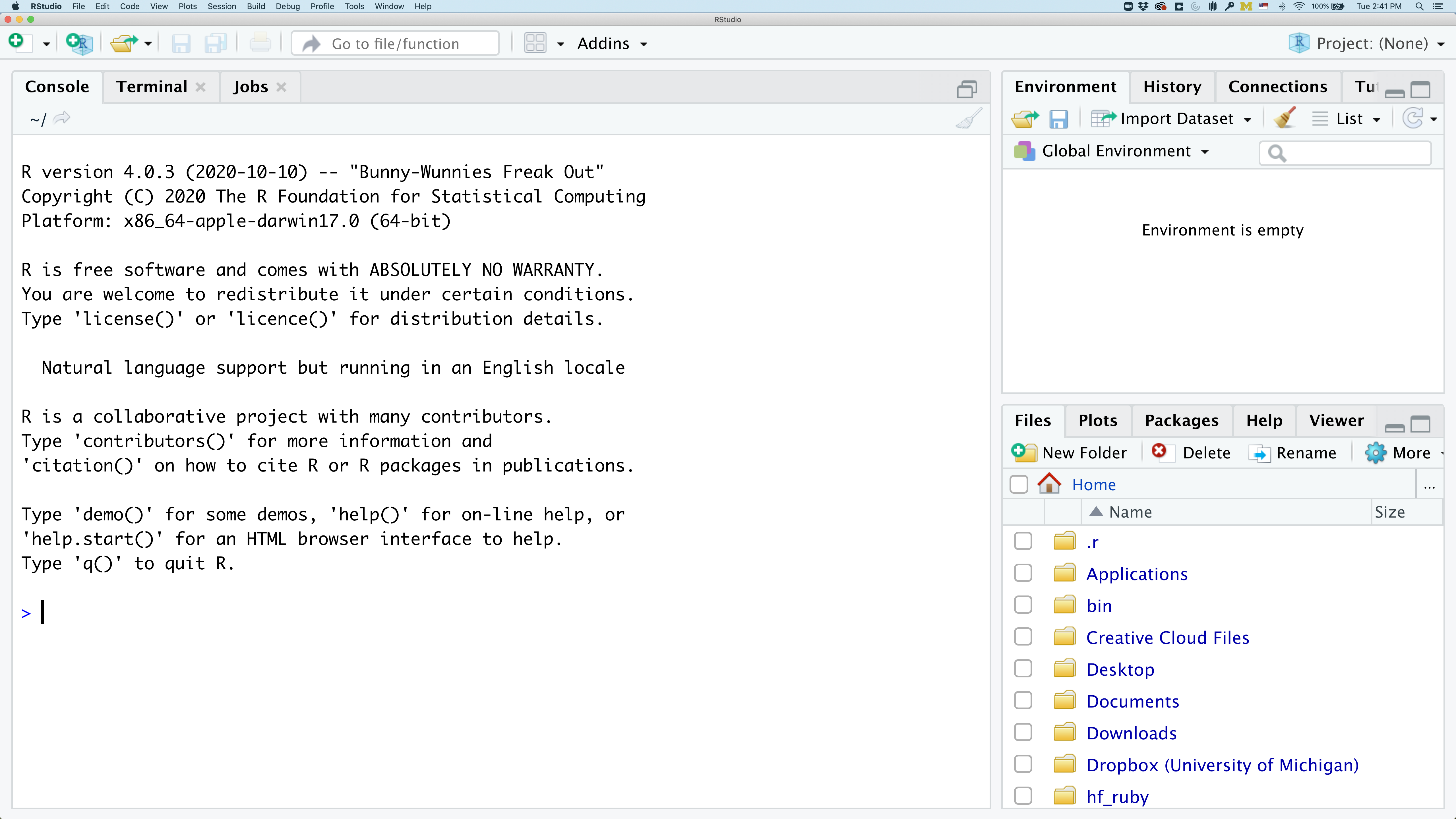
On the left you’ll have a panel with three tabs - Console, Terminal, and Jobs. The Console tab is what running R from the command line looks like. This is where you can enter R code. Try typing in 2+2 at the prompt (>). In the upper right panel are tabs indicating the Environment, History, and a few other things. If you click on the History tab, you’ll see the command you ran at the R prompt.
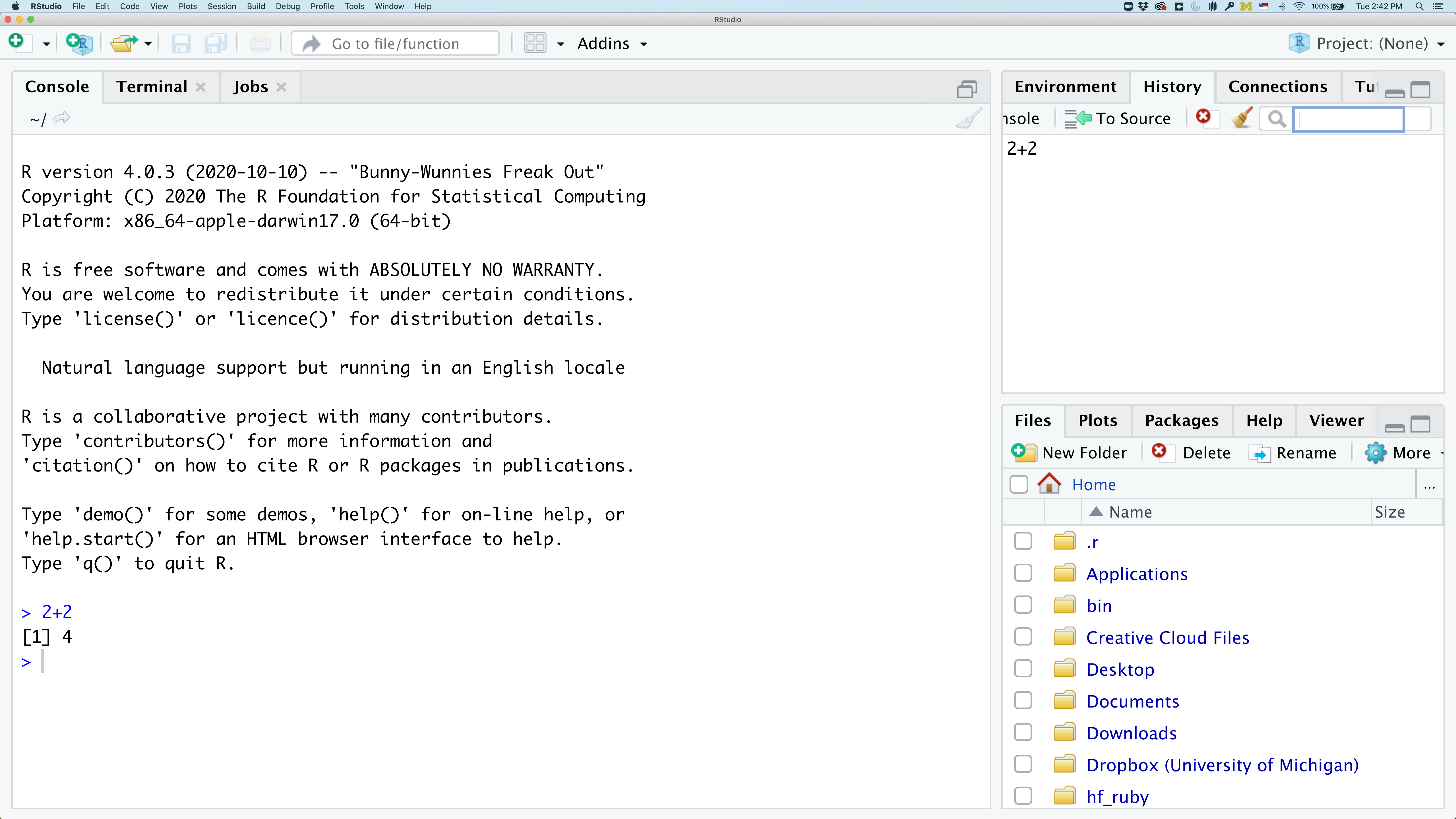
In the lower right panel are tabs for Files, Plots, Packages, Help, and Viewer. You used the Packages tab to install tidyverse.
We’ll spend more time in each of these tabs as we go through the workshop, so we won’t spend a lot of time discussing them now.
You might want to alter the appearance of your RStudio window. The default appearance has a white background with black text. If you go to the Tools menu at the top of your screen, you’ll see a “Global options” menu at the bottom of the drop down; select that.
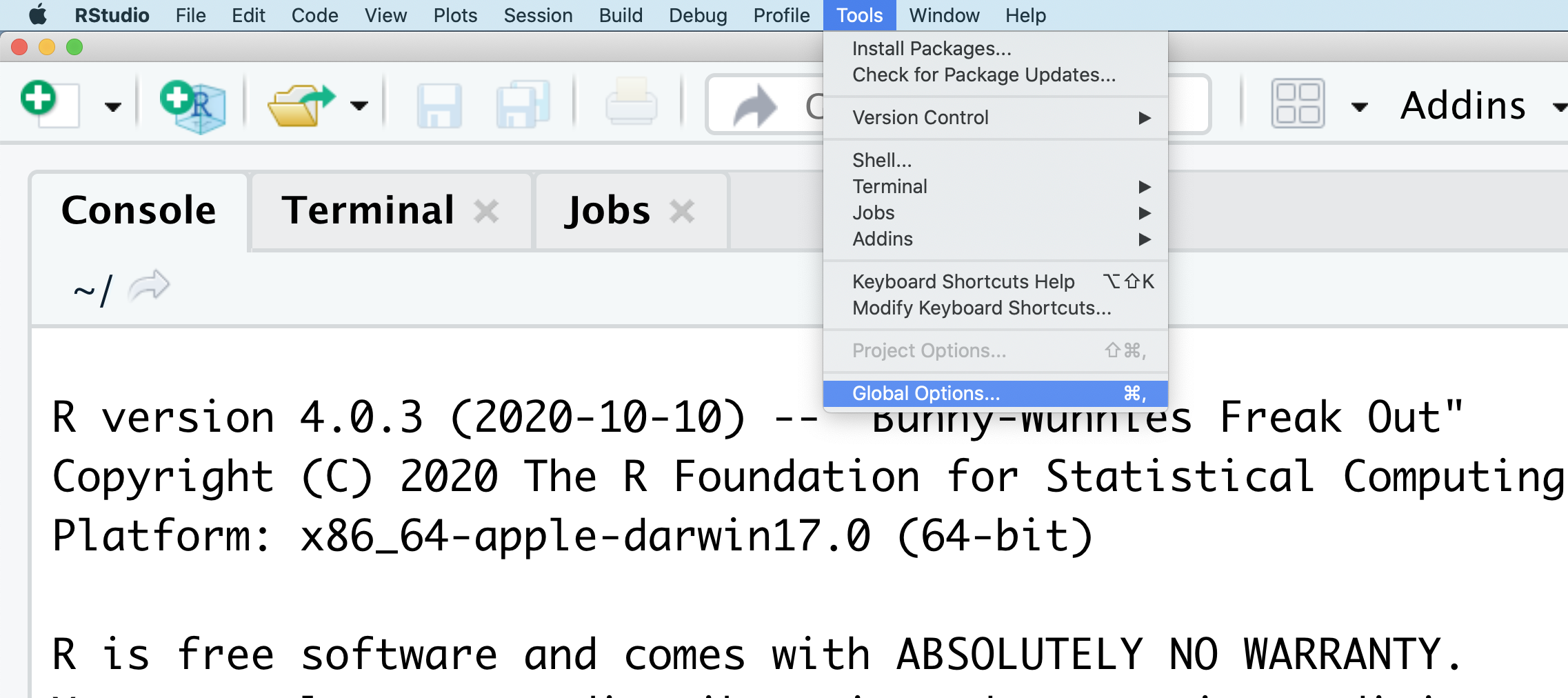
From there you will see the ability to alter numerous things about RStudio. Under the Appearances tab you can select the theme you like most. As you can see there’s a lot in Global options that you can set to improve your experience in RStudio. Most of these settings are a matter of personal preference.
However, you can update settings to help you to insure the reproducibility of your code. In the General tab, none of the selectors in the R Sessions, Workspace, and History should be selected. In addition, the toggle next to “Save workspace to .RData on exit” should be set to never. These setting will help ensure that things you worked on previously don’t carry over between sessions.
Let’s get going on our analysis!
One of the helpful features in RStudio is the ability to create a project. A project is a special directory that contains all of the code and data that you will need to run an analysis.
At the top of your screen you’ll see the “File” menu. Select that menu and then the menu for “New Project…”.
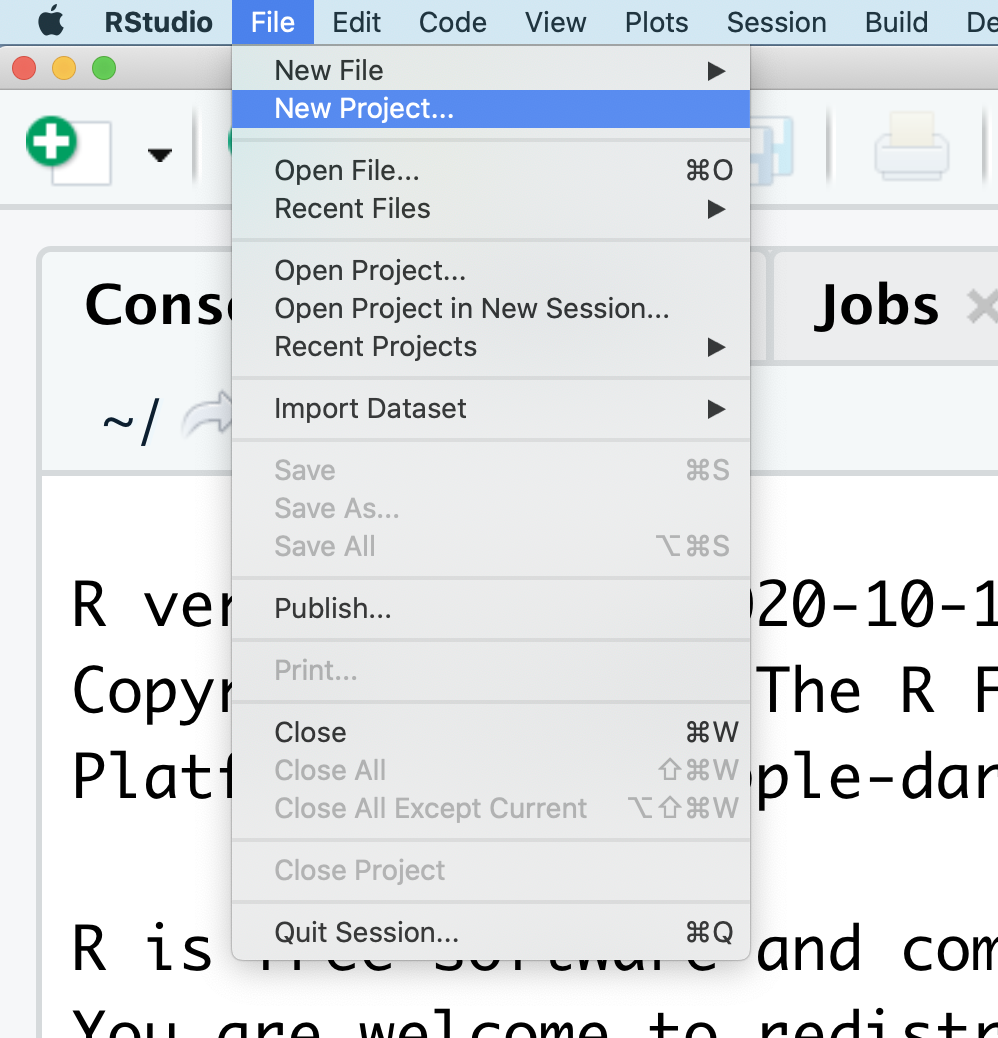
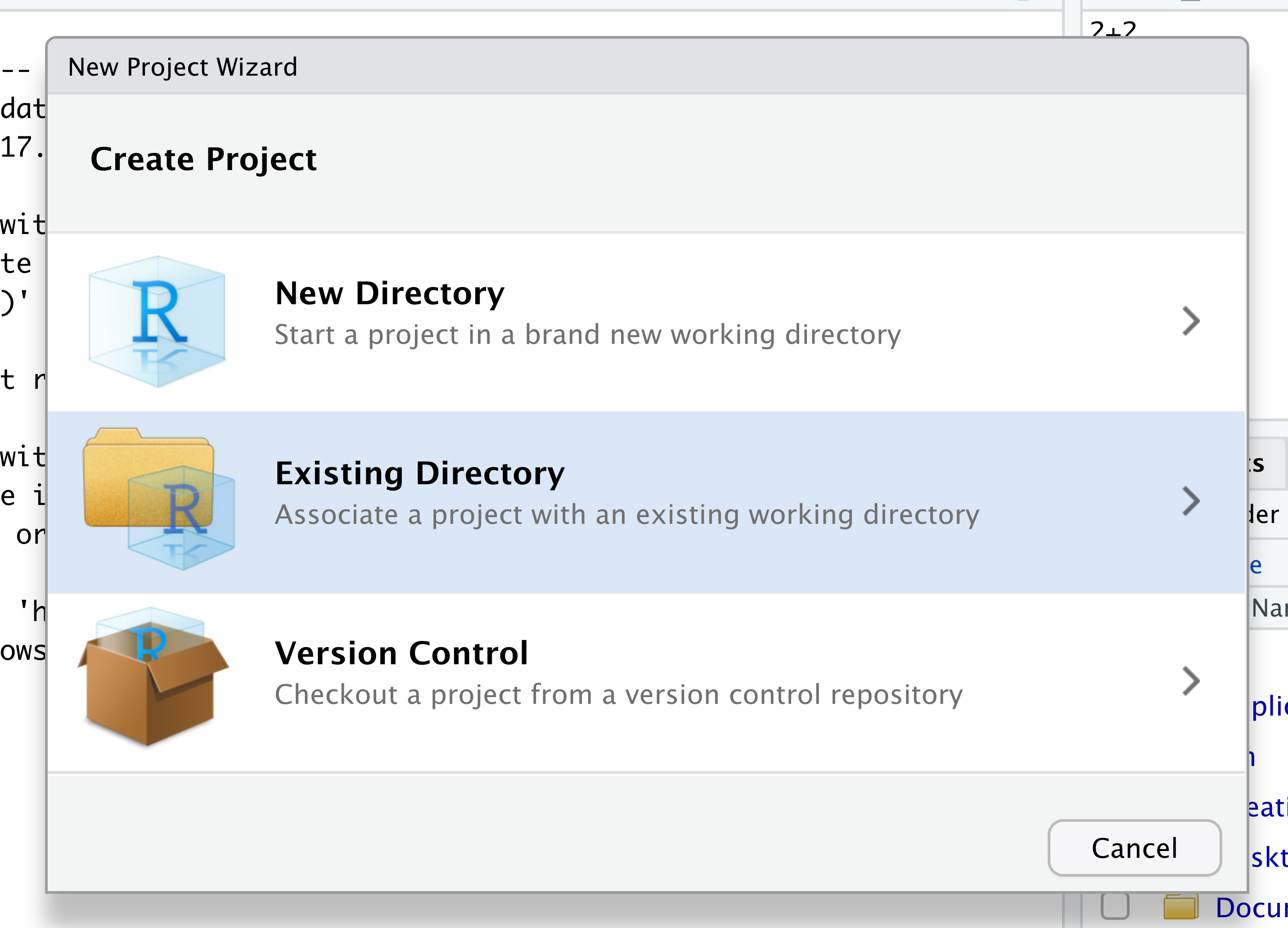
When the smaller window opens, select “Existing Directory” and then the “Browse” button in the next window.
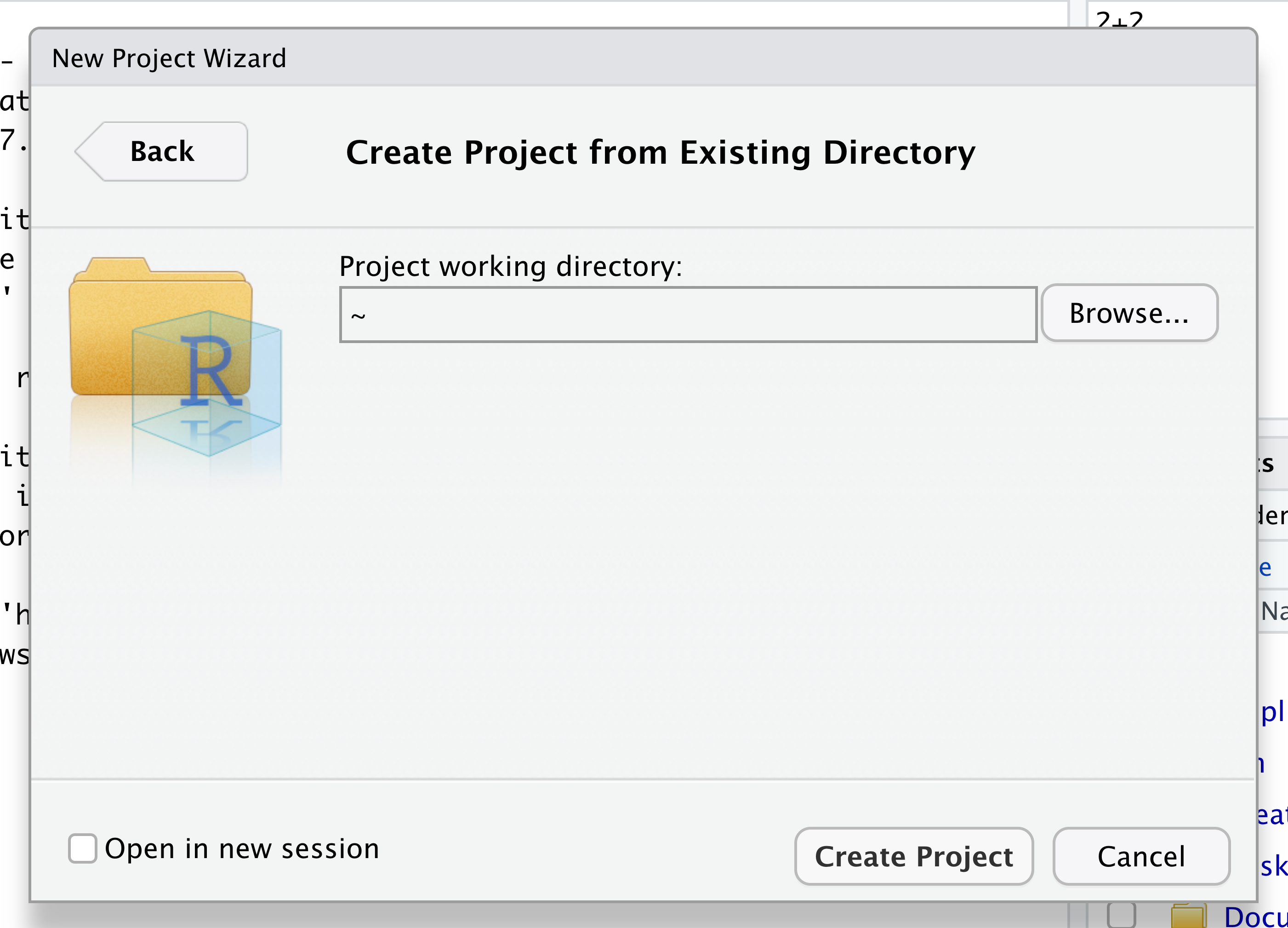
Navigate to the directory that contains your code and data from the setup instructions and click the “Open” button.
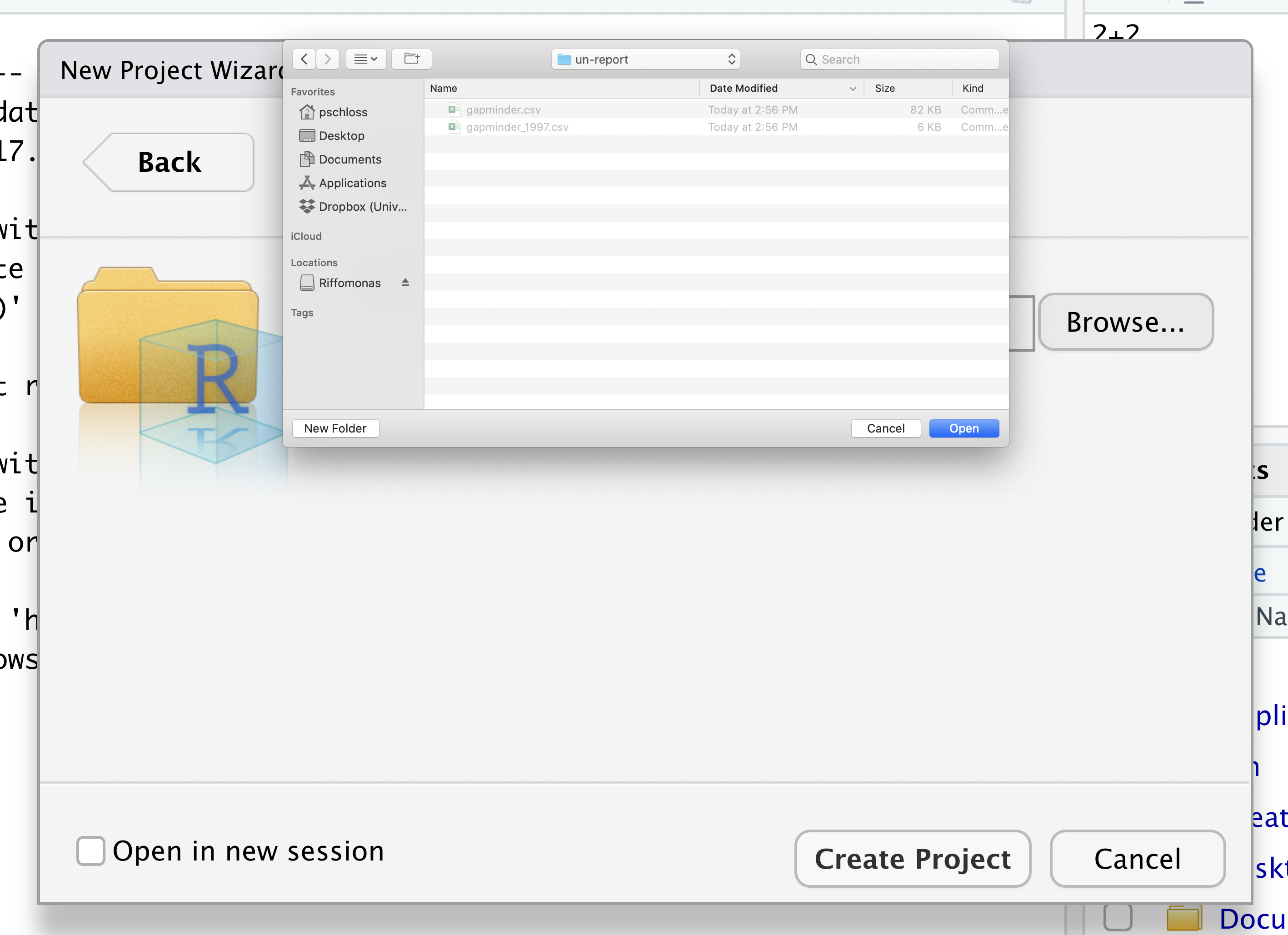
Then click the “Create Project” button.
Did you notice anything change?
In the lower right corner of your RStudio session, you should notice that your Files tab is now your project directory. You’ll also see a file called un-report.Rproj in that directory.
From now on, you should start RStudio by double clicking on that file. This will make sure you are in the correct directory when you run your analysis.
We’d like to create a file where we can keep track of our R code.
Back in the “File” menu, you’ll see the first option is “New File”. Selecting “New File” opens another menu to the right and the first option is “R Script”. Select “R Script”.
Now we have a fourth panel in the upper left corner of RStudio that includes an Editor tab with an untitled R Script. Let’s save this file as gdp_population.R in our project directory.
We will be entering R code into the Editor tab to run in our Console panel.
On line 1 of gdp_population.R, type 2+2.
With your cursor on the line with the 2+2, click the button that says Run. You should be able to see that 2+2 was run in the Console.
As you write more code, you can highlight multiple lines and then click Run to run all of the lines you have selected.
Let’s delete the line with 2+2 and replace it with library(tidyverse).
Go ahead and run that line in the Console by clicking the Run button on the top right of the Editor tab and choosing Run Selected Lines. This loads a set of useful functions and sample data that makes it easier for us to do complex analyses and create professional visualizations in R.
library(tidyverse)
── Attaching packages ───────────────────────────────────────────────────────────────────────────────────────────────────────────────────── tidyverse 1.3.1 ──
✔ ggplot2 3.3.5 ✔ purrr 0.3.4
✔ tibble 3.1.5 ✔ dplyr 1.0.7
✔ tidyr 1.1.4 ✔ stringr 1.4.0
✔ readr 2.0.2 ✔ forcats 0.5.1
── Conflicts ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
What’s with all those messages???
When you loaded the
tidyversepackage, you probably got a message like the one we got above. Don’t panic! These messages are just giving you more information about what happened when you loadedtidyverse.Tidyverseis actually a collection of several different packages, so the first section of the message tells us what packages were installed when we loadedtidyverse(these includeggplot2, which we’ll be using a lot in this lesson, anddyplr, which you’ll be introduced to tomorrow in the R for Data Analysis lesson).
The second section of messages gives a list of “conflicts.” Sometimes, the same function name will be used in two different packages, and R has to decide which function to use. For example, our message says that:
dplyr::filter() masks stats::filter()This means that two different packages (
dyplrfromtidyverseandstatsfrom base R) have a function namedfilter(). By default, R uses the function that was most recently loaded, so if we try using thefilter()function after loadingtidyverse, we will be using thefilter()function fromdplyr().
Hint / Pro-tip
Those of us that use R on a daily basis use cheat sheets to help us remember how to use various R functions. If you haven’t already, print out the PDF versions of the cheat sheets that were in the setup instructions.
You can also find them in RStudio by going to the “Help” menu and selecting “Cheat Sheets”. The two that will be most helpful in this workshop are “Data Visualization with ggplot2”, “Data Transformation with dplyr”, “R Markdown Cheat Sheet”, and “R Markdown Reference Guide”.
For things that aren’t on the cheat sheets, Google is your best friend. Even expert coders use Google when they’re stuck or trying something new!
Loading and reviewing data
We will import a subsetted file from the gapminder dataset called gapminder_1997.csv. There are many ways to import data into R but for your first plot we will use RStudio’s file menu to import and display this data. As we move through this process, RStudio will translate these point and click commands into code for us.
In RStudio select “File” > “Import Dataset” > “From Text (readr)”.
The file is located in the main directory, click the “Browse” button and select the file named gapminder_1997.csv. A preview of the data will appear in the window. You can see there are a lot of Import Options listed, but R has chosen the correct defaults for this particular file.
We can see in that box that our data will be imported with the Name: “gapminder_1997”. Also note that this screen will show you all the code that will be run when you import your data in the lower right “Code Preview”. Since everything looks good, click the “Import” button to bring your data into R.
After you’ve imported your data, a table will open in a new tab in the top left corner of RStudio. This is a quick way to browse your data to make sure everything looks like it has been imported correctly. To review the data, click on the new tab.
We see that our data has 5 columns (variables).
Each row contains life expectancy (“lifeExp”), the total population (“pop”), and the per capita gross domestic product (“gdpPercap”) for a given country (“country”).
There is also a column that says which continent each country is in (“continent”). Note that both North America and South America are combined into one category called “Americas”.
After we’ve reviewed the data, you’ll want to make sure to click the tab in the upper left to return to your gdp_population.R file so we can start writing some code.
Now look in the Environment tab in the upper right corner of RStudio. Here you will see a list of all the objects you’ve created or imported during your R session. You will now see gapminder_1997 listed here as well.
Finally, take a look at the Console at the bottom left part of the RStudio screen. Here you will see the commands that were run for you to import your data in addition to associated metadata and warnings.
Data objects
There are many different ways to store data in R. Most objects have a table-like structure with rows and columns. We will refer to these objects generally as “data objects”. If you’ve used R before, you many be used to calling them “data.frames”. Functions from the “tidyverse” such as
read_csvwork with objects called “tibbles”, which are a specialized kind of “data.frame.” Another common way to store data is a “data.table”. All of these types of data objects (tibbles, data.frames, and data.tables) can be used with the commands we will learn in this lesson to make plots. We may sometimes use these terms interchangeably.
Understanding commands
Let’s start by looking at the code RStudio ran for us by copying and pasting the first line from the console into our gdp_population.R file that is open in the Editor window.
gapminder_1997 <- read_csv("gapminder_1997.csv")
Rows: 142 Columns: 5
── Column specification ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
Delimiter: ","
chr (2): country, continent
dbl (3): pop, lifeExp, gdpPercap
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
You should now have a line of text in your code file that started with gapminder and ends with a ) symbol.
What if we want to run this command from our code file?
In order to run code that you’ve typed in the editor, you have a few options. We can click Run again from the right side of the Editor tab but the quickest way to run the code is by pressing Ctrl+Enter on your keyboard.
This will run the line of code that currently contains your cursor and will move your cursor to the next line. Note that when Rstudio runs your code, it basically just copies your code from the Editor window to the Console window, just like what happened when we selected Run Selected Line(s).
Let’s take a closer look at the parts of this command.
Starting from the left, the first thing we see is gapminder_1997. We viewed the contents of this file after it was imported so we know that gapminder_1997 acts as a placeholder for our data.
If we highlight just gapminder_1997 within our code file and press Ctrl+Enter on our keyboard, what do we see?
We should see a data table outputted, similar to what we saw in the Viewer tab.
In R terms, gapminder_1997 is a named object that references or stores something. In this case, gapminder_1997 stores a specific table of data.
Looking back at the command in our code file, the second thing we see is a <- symbol, which is the assignment operator. It assigns values generated or typed on the right to objects on the left. An alternative symbol that you might see used as an assignment operator is the = but it is clearer to only use <- for assignment. We use this symbol so often that RStudio has a keyboard short cut for it: Alt+- on Windows, and Option+- on Mac.
Exercise: Assigning values to objects
Try to assign values to some objects and observe each object after you have assigned a new value. What do you notice?
name <- "Ben" name age <- 26 age name <- "Harry Potter" nameSolution
When we assign a value to an object, the object stores that value so we can access it later. However, if we store a new value in an object we have already created (like when we stored “Harry Potter” in the
nameobject), it replaces the old value. Theageobject does not change, because we never assign it a new value.
Guidelines on naming objects
- You want your object names to be explicit and not too long.
- They cannot start with a number (2x is not valid, but x2 is).
- R is case sensitive, so for example, weight_kg is different from Weight_kg.
- You cannot use spaces in the name.
- There are some names that cannot be used because they are the names of fundamental functions in R (e.g., if, else, for; see here for a complete list). If in doubt, check the help to see if the name is already in use (
?function_name).- It’s best to avoid dots (.) within names. Many function names in R itself have them and dots also have a special meaning (methods) in R and other programming languages.
- It is recommended to use nouns for object names and verbs for function names.
- Be consistent in the styling of your code, such as where you put spaces, how you name objects, etc. Using a consistent coding style makes your code clearer to read for your future self and your collaborators. One popular style guide can be found through the tidyverse.
Bonus Exercise: Bad names for objects
Try to assign values to some new objects. What do you notice? After running all four lines of code bellow, what value do you think the object
Flowerholds?1number <- 3 Flower <- "marigold" flower <- "rose" favorite number <- 12Solution
Notice that we get an error when we try to assign values to
1numberandfavorite number. This is because we cannot start an object name with a numeral and we cannot have spaces in object names. The objectFlowerstill holds “marigold.” This is because R is case-sensitive, so runningflower <- "rose"does NOT change theFlowerobject. This can get confusing, and is why we generally avoid having objects with the same name and different capitalization.
The next part of the command is read_csv("gapminder_1997.csv"). This has a few different key parts. The first part is the read_csv function. You call a function in R by typing it’s name followed by opening then closing parenthesis. Each function has a purpose, which is often hinted at by the name of the function. Let’s try to run the function without anything inside the parenthesis.
read_csv()
Error in vroom::vroom(file, delim = ",", col_names = col_names, col_types = col_types, : argument "file" is missing, with no default
We get an error message. Don’t panic! Error messages pop up all the time, and can be super helpful in debugging code.
In this case, the message tells us “argument “file” is missing, with no default.” Many functions, including read_csv, require additional pieces of information to do their job. We call these additional values “arguments” or “parameters.” You pass arguments to a function by placing values in between the parenthesis. A function takes in these arguments and does a bunch of “magic” behind the scenes to output something we’re interested in.
For example, when we loaded in our data, the command contained "gapminder_1997.csv" inside the read_csv() function. This is the value we assigned to the file argument. But we didn’t say that that was the file. How does that work?
Hint/Pro-tip
Each function has a help page that documents what arguments the function expects and what value it will return. You can bring up the help page a few different ways. If you have typed the function name in the Editor windows, you can put your cursor on the function name and press F1 to open help page in the Help viewer in the lower right corner of RStudio. You can also type
?followed by the function name in the console. For example, try running?read_csv. A help page should pop up with information about what the function is used for and how to use it, as well as useful examples of the function in action. As you can see, the first argument ofread_csvis the file path.
The read_csv() function took the file path we provided, did who-knows-what behind the scenes, and then outputted an R object with the data stored in that csv file. All that, with one short line of code!
Do all functions need arguments? Let’s test some other functions:
Sys.Date()
[1] "2021-10-22"
getwd()
[1] "/Users/cgates/git/temp/_episodes_rmd"
Comments
Sometimes you may want to write comments in your code to help you remember what your code is doing, but you don’t want R to think these comments are a part of the code you want to evaluate. That’s where comments come in! Anything after a
#symbol in your code will be ignored by R. For example, let’s say we wanted to make a note of what each of the functions we just used do:Sys.Date() # outputs the current date[1] "2021-10-22"getwd() # outputs our current working directory (folder)[1] "/Users/cgates/git/temp/_episodes_rmd"
While some functions, like those above, don’t need any arguments, in other functions we may want to use multiple arguments. When we’re using multiple arguments, we separate the arguments with commas. For example, we can use the sum() function to add numbers together:
sum(5, 6)
[1] 11
Exercise: Learning more about functions
Look up the function
round. What does it do? What will you get as output for the following lines of code?round(3.1415) round(3.1415,3)Solution
roundrounds a number. By default, it rounds it to zero digits (in our example above, to 3). If you give it a second number, it rounds it to that number of digits (in our example above, to 3.141)
Notice how in this example, we didn’t include any argument names. But you can use argument names if you want:
read_csv(file = 'gapminder_1997.csv')
Rows: 142 Columns: 5
── Column specification ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
Delimiter: ","
chr (2): country, continent
dbl (3): pop, lifeExp, gdpPercap
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
# A tibble: 142 × 5
country pop continent lifeExp gdpPercap
<chr> <dbl> <chr> <dbl> <dbl>
1 Afghanistan 22227415 Asia 41.8 635.
2 Albania 3428038 Europe 73.0 3193.
3 Algeria 29072015 Africa 69.2 4797.
4 Angola 9875024 Africa 41.0 2277.
5 Argentina 36203463 Americas 73.3 10967.
6 Australia 18565243 Oceania 78.8 26998.
7 Austria 8069876 Europe 77.5 29096.
8 Bahrain 598561 Asia 73.9 20292.
9 Bangladesh 123315288 Asia 59.4 973.
10 Belgium 10199787 Europe 77.5 27561.
# … with 132 more rows
Exercise: Position of the arguments in functions
Which of the following lines of code will give you an output of 3.14? For the one(s) that don’t give you 3.14, what do they give you?
round(x = 3.1415) round(x = 3.1415, digits = 2) round(digits = 2, x = 3.1415) round(2, 3.1415)Solution
The 2nd and 3rd lines will give you the right answer because the arguments are named, and when you use names the order doesn’t matter. The 1st line will give you 3 because the default number of digits is 0. Then 4th line will give you 2 because, since you didn’t name the arguments, x=2 and digits=3.1415.
Sometimes it is helpful - or even necessary - to include the argument name, but often we can skip the argument name, if the argument values are passed in a certain order. If all this function stuff sounds confusing, don’t worry! We’ll see a bunch of examples as we go that will make things clearer.
Exercise: Reading in an excel file
Say you have an excel file and not a csv - how would you read that in? Hint: Use the Internet to help you figure it out!
Solution
One way is using the
read_excelfunction in thereadxlpackage. There are other ways, but this is our preferred method because the output will be the same as the output ofread_csv.
Creating our first plot
We will be using the ggplot2 package today to make our plots. This is a very powerful package that creates professional looking plots and is one of the reasons people like using R so much. All plots made using the ggplot2 package start by calling the ggplot() function. So in the tab you created for the gdp_population.R file, type the following:
ggplot(data=gapminder_1997)

To run code that you’ve typed in the editor, you have a few options. Remember that the quickest way to run the code is by pressing Ctrl+Enter on your keyboard. This will run the line of code that currently contains your cursor or any highlighted code.
When we run this code, the Plots tab will pop to the front in the lower right corner of the RStudio screen. Right now, we just see a big grey rectangle.
What we’ve done is created a ggplot object and told it we will be using the data from the gapminder_1997 object that we’ve loaded into R. We’ve done this by calling the ggplot() function with gapminder_1997 as the data argument.
So we’ve made a plot object, now we need to start telling it what we actually want to draw in this plot. The elements of a plot have a bunch of properties like an x and y position, a size, a color, etc. These properties are called aesthetics. When creating a data visualization, we map a variable in our dataset to an aesthetic in our plot. In ggplot, we can do this by creating an “aesthetic mapping”, which we do with the aes() function.
To create our plot, we need to map variables from our gapminder_1997 object to ggplot aesthetics using the aes() function. Since we have already told ggplot that we are using the data in the gapminder_1997 object, we can access the columns of gapminder_1997 using the object’s column names. (Remember, R is case-sensitive, so we have to be careful to match the column names exactly!)
We are interested in whether there is a relationship between GDP and life expectancy, so let’s start by telling our plot object that we want to map our GDP values to the x axis of our plot. We do this by adding (+) information to our plot object. Add this new line to your code and run both lines by highlighting them and pressing Ctrl+Enter on your keyboard:
ggplot(data = gapminder_1997) +
aes(x = gdpPercap)
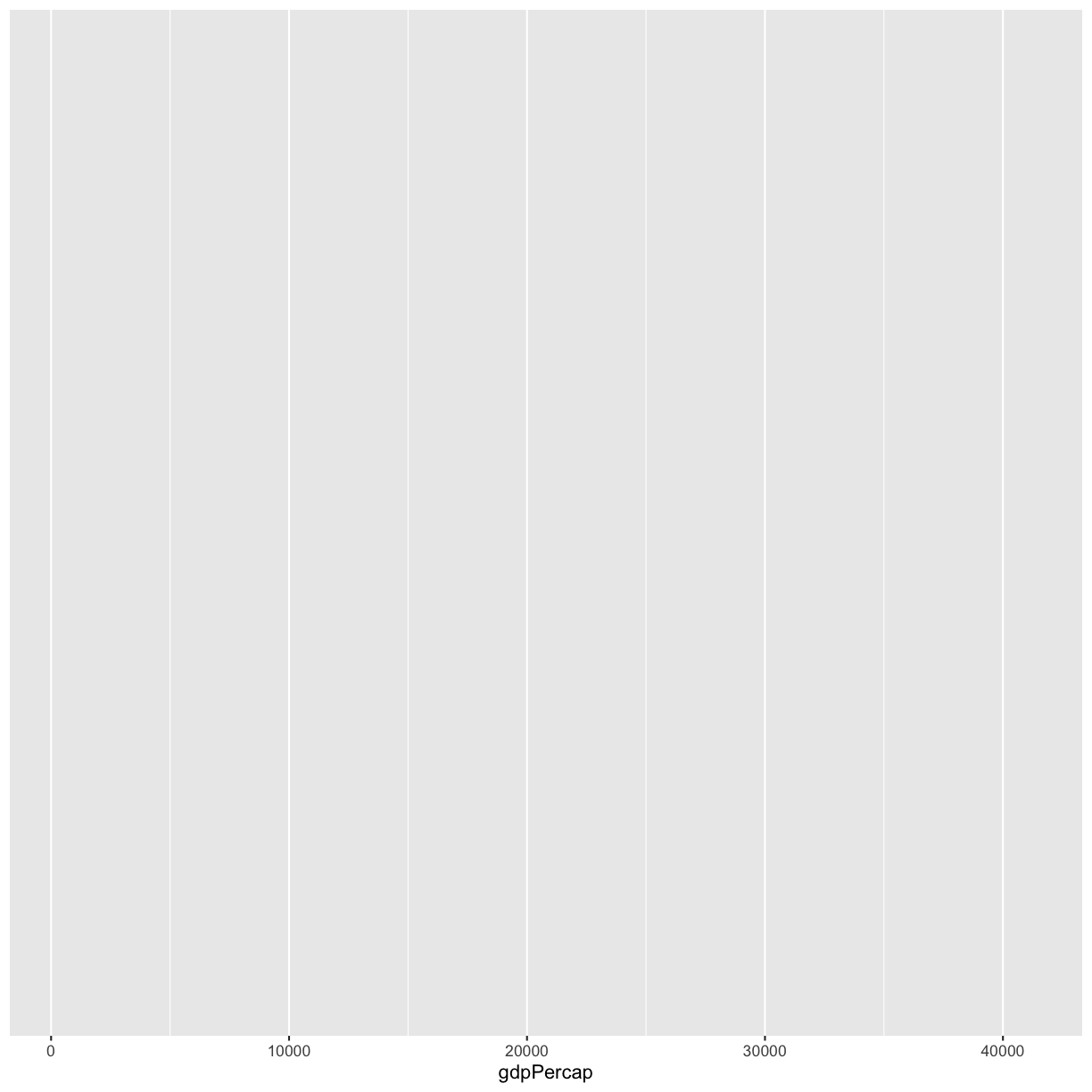
Note that we’ve added this new function call to a second line just to make it easier to read. To do this we make sure that the + is at the end of the first line otherwise R will assume your command ends when it starts the next row. The + sign indicates not only that we are adding information, but to continue on to the next line of code.
Observe that our Plot window is no longer a grey square. We now see that we’ve mapped the gdpPercap column to the x axis of our plot. Note that that column name isn’t very pretty as an x-axis label, so let’s add the labs() function to make a nicer label for the x axis
ggplot(data = gapminder_1997) +
aes(x = gdpPercap) +
labs(x = "GDP Per Capita")
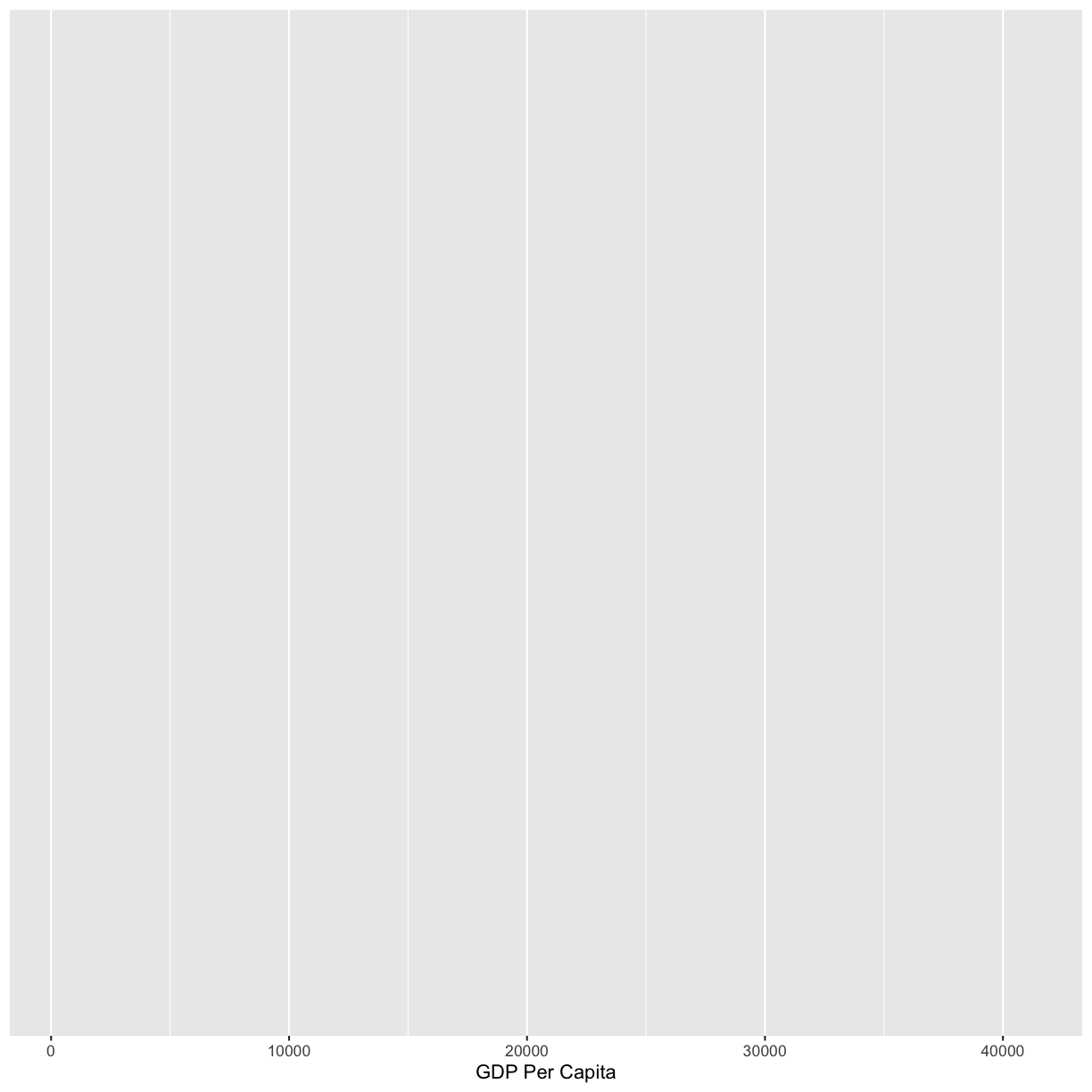
OK. That looks better.
Quotes vs No Quotes
Notice that when we added the label value we did so by placing the values inside quotes. This is because we are not using a value from inside our data object - we are providing the name directly. When you need to include actual text values in R, they will be placed inside quotes to tell them apart from other object or variable names. The general rule is that if you want to use values from the columns of your data object, then you supply the name of the column without quotes, but if you want to specify a value that does not come from your data, then use quotes.
Exercise: Mapping life expectancy to the y axis
Map our
lifeExpvalues to the y axis and give them a nice label.Solution
ggplot(data = gapminder_1997) + aes(x = gdpPercap) + labs(x = "GDP Per Capita") + aes(y = lifeExp) + labs(y = "Life Expectancy")
Excellent. We’ve now told our plot object where the x and y values are coming from and what they stand for. But we haven’t told our object how we want it to draw the data. There are many different plot types (bar charts, scatter plots, histograms, etc). We tell our plot object what to draw by adding a “geometry” (“geom” for short) to our object. We will talk about many different geometries today, but for our first plot, let’s draw our data using the “points” geometry for each value in the data set. To do this, we add geom_point() to our plot object:
ggplot(data = gapminder_1997) +
aes(x = gdpPercap) +
labs(x = "GDP Per Capita") +
aes(y = lifeExp) +
labs(y = "Life Expectancy") +
geom_point()
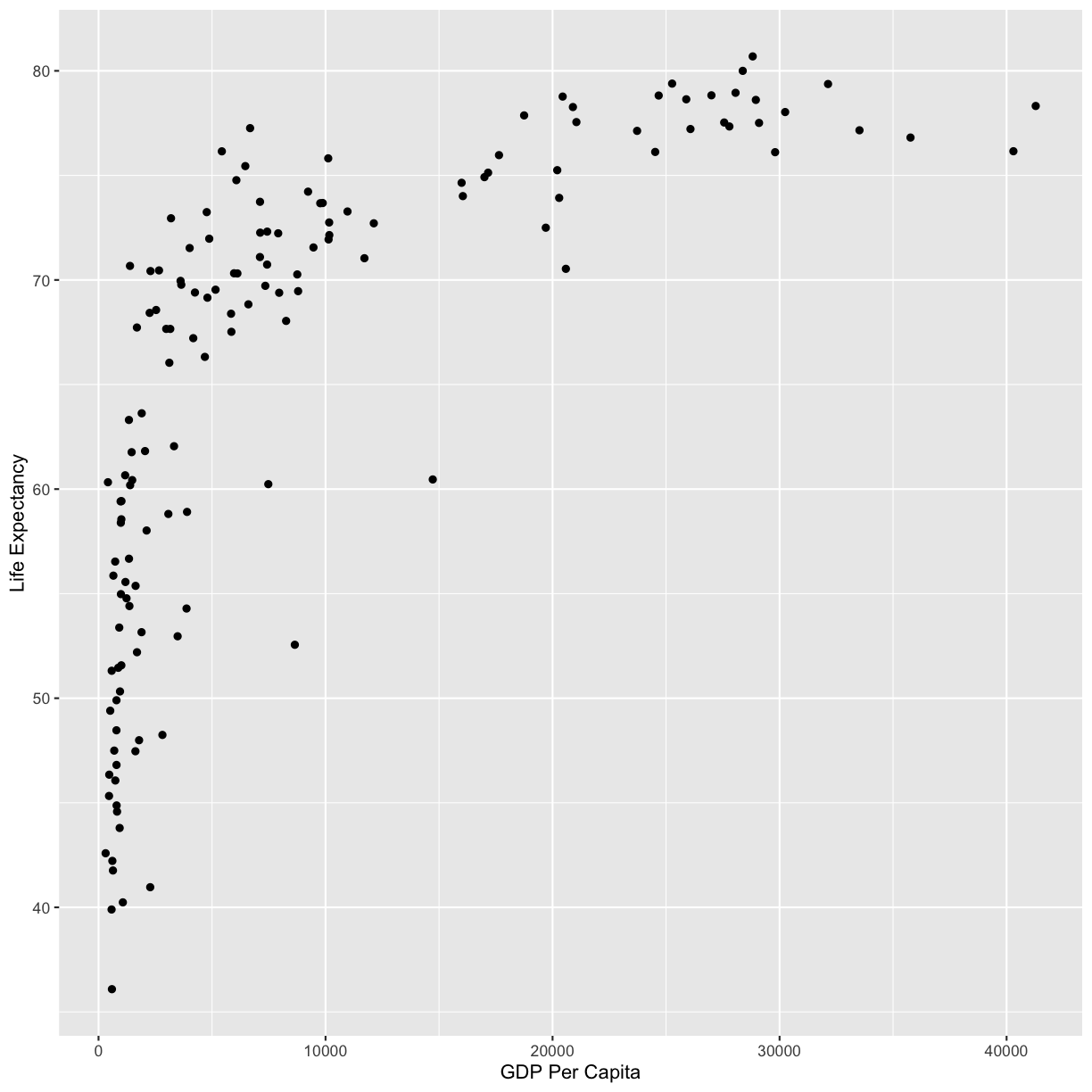
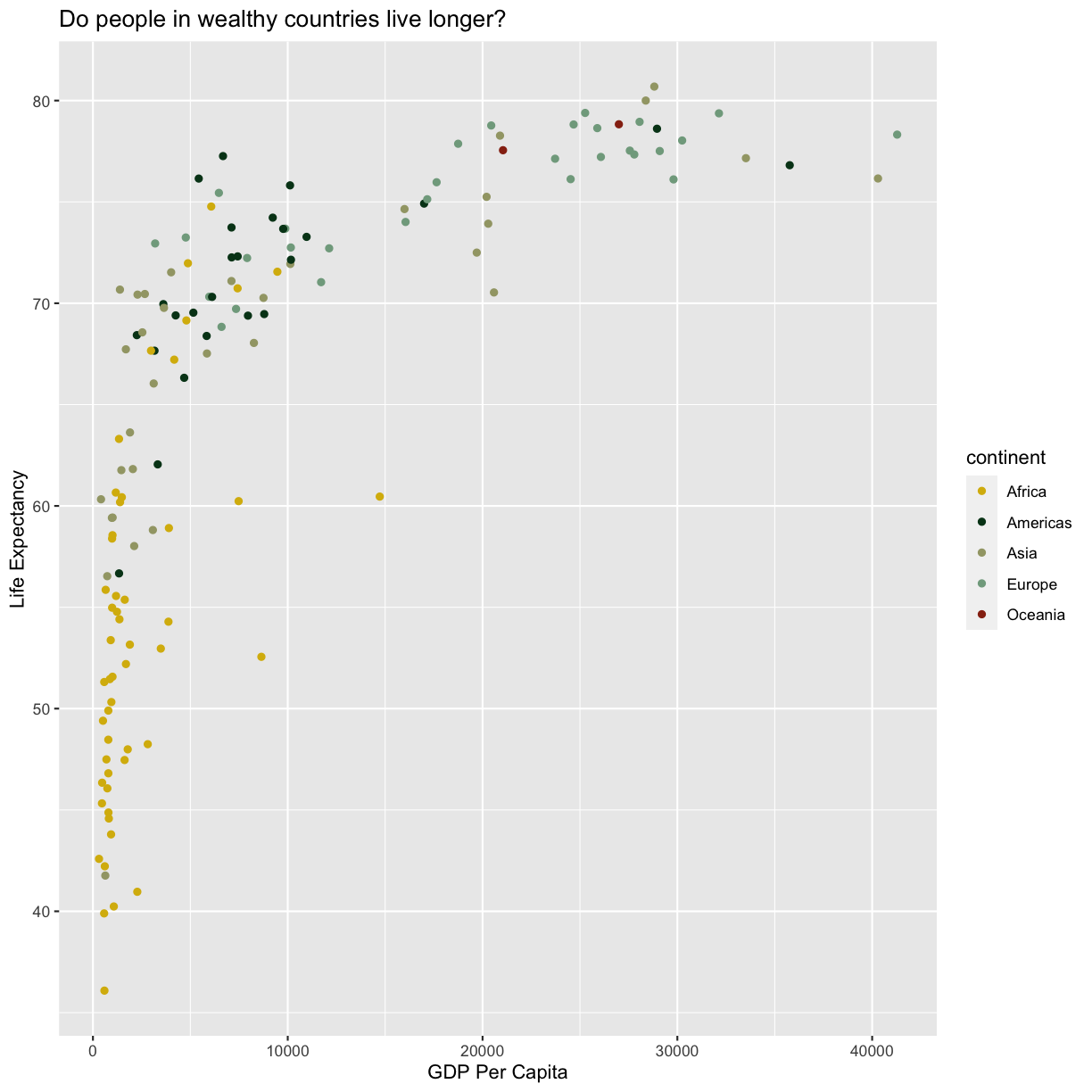
Now we’re really getting somewhere. It finally looks like a proper plot! We can now see a trend in the data. It looks like countries with a larger GDP tend to have a higher life expectancy. Let’s add a title to our plot to make that clearer. Again, we will use the labs() function, but this time we will use the title = argument.
ggplot(data = gapminder_1997) +
aes(x = gdpPercap) +
labs(x = "GDP Per Capita") +
aes(y = lifeExp) +
labs(y = "Life Expectancy") +
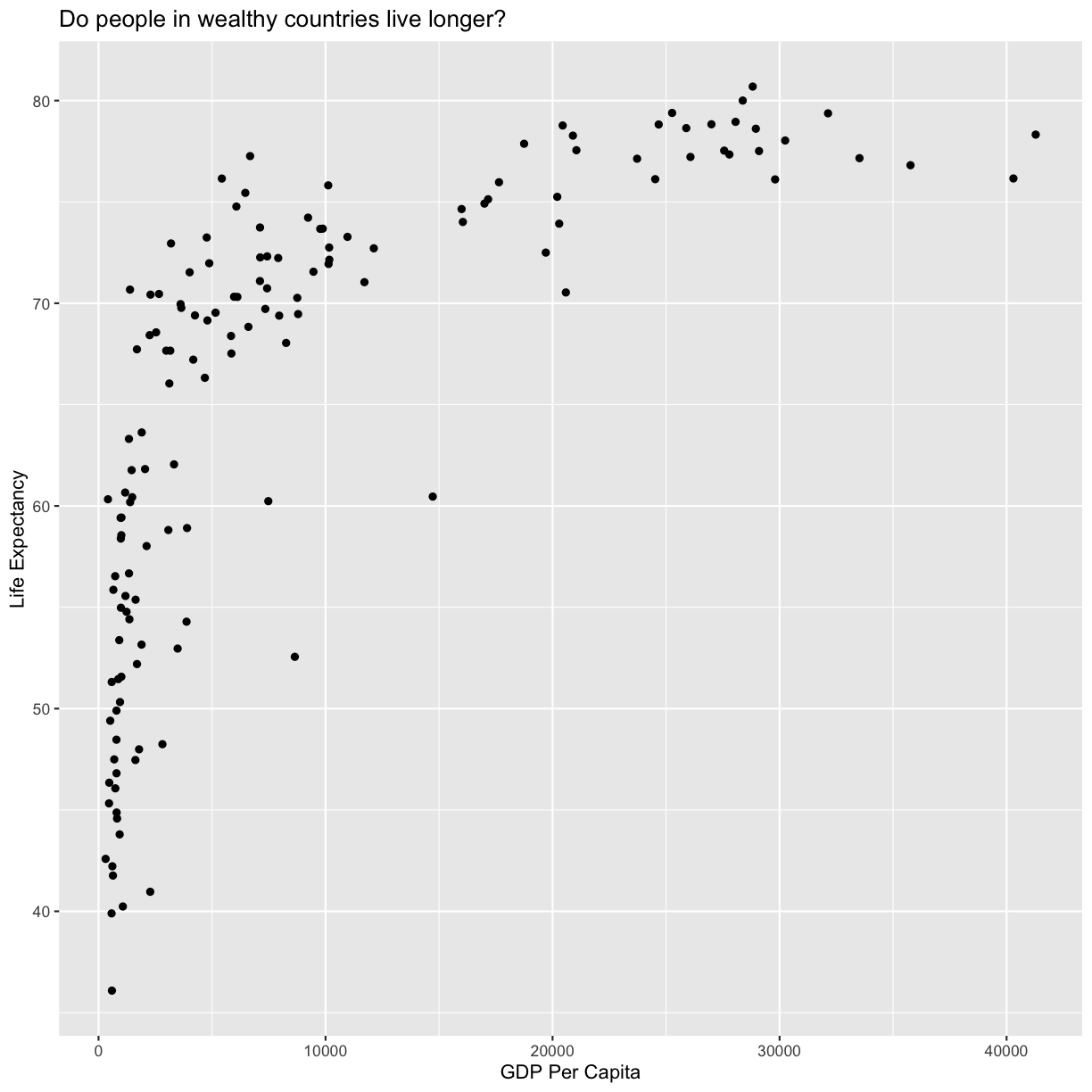
geom_point() +
labs(title = "Do people in wealthy countries live longer?")
No one can deny we’ve made a very handsome plot! But now looking at the data, we might be curious about learning more about the points that are the extremes of the data. We know that we have two more pieces of data in the gapminder_1997 object that we haven’t used yet. Maybe we are curious if the different continents show different patterns in GDP and life expectancy. One thing we could do is use a different color for each of the continents. To map the continent of each point to a color, we will again use the aes() function:
ggplot(data = gapminder_1997) +
aes(x = gdpPercap) +
labs(x = "GDP Per Capita") +
aes(y = lifeExp) +
labs(y = "Life Expectancy") +
geom_point() +
labs(title = "Do people in wealthy countries live longer?") +
aes(color = continent)
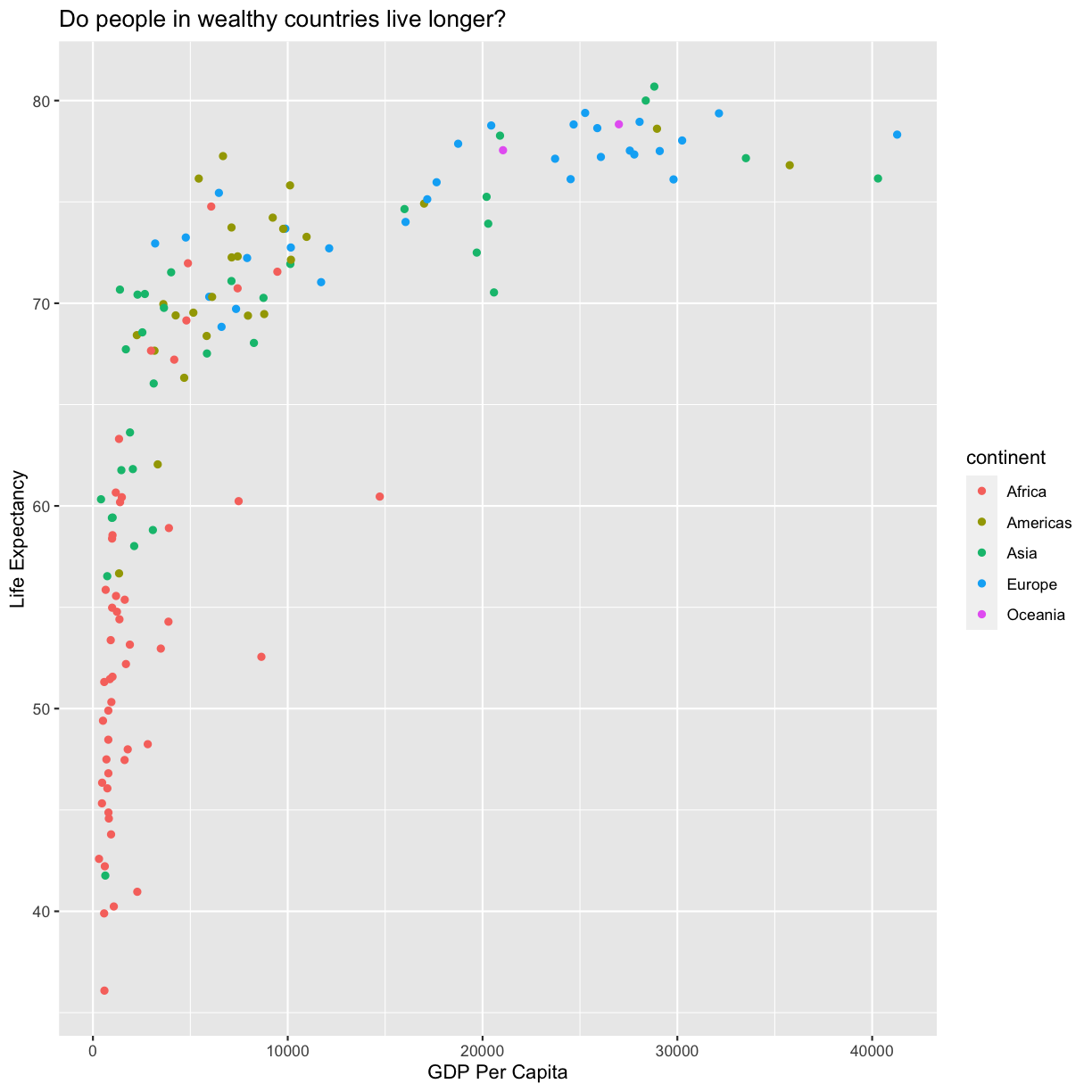
Here we can see that in 1997 the African countries had much lower life expectancy than many other continents. Notice that when we add a mapping for color, ggplot automatically provided a legend for us. It took care of assigning different colors to each of our unique values of the continent variable. (Note that when we mapped the x and y values, those drew the actual axis labels, so in a way the axes are like the legends for the x and y values). The colors that ggplot uses are determined by the color “scale”. Each aesthetic value we can supply (x, y, color, etc) has a corresponding scale. Let’s change the colors to make them a bit prettier.
ggplot(data = gapminder_1997) +
aes(x = gdpPercap) +
labs(x = "GDP Per Capita") +
aes(y = lifeExp) +
labs(y = "Life Expectancy") +
geom_point() +
labs(title = "Do people in wealthy countries live longer?") +
aes(color = continent) +
scale_color_brewer(palette = "Set1")
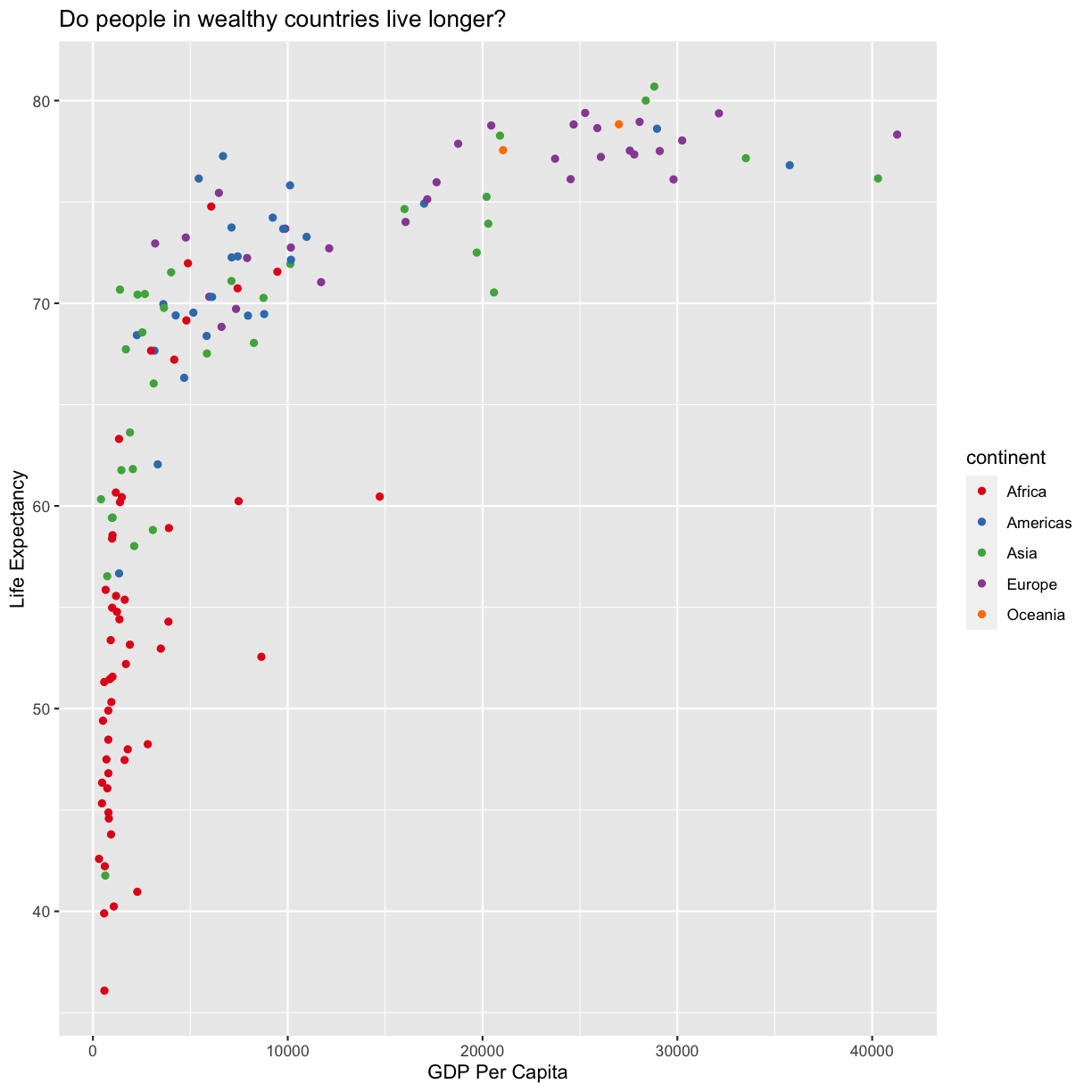
The scale_color_brewer() function is just one of many you can use to change colors. There are bunch of “palettes” that are build in. You can view them all by running RColorBrewer::display.brewer.all() or check out the Color Brewer website for more info about choosing plot colors.
There are also lots of other fun options:
Bonus Exercise: Changing colors
Play around with different color palettes. Feel free to install another package and choose one of those if you want. Pick your favorite!
Solution
You can use RColorBrewer::display.brewer.all() to pick a color palette. As a bonus, you can also use one of the packages listed above. Here’s an example:
#install.packages("wesanderson") # install package from GitHub library(wesanderson) ggplot(data = gapminder_1997) + aes(x = gdpPercap) + labs(x = "GDP Per Capita") + aes(y = lifeExp) + labs(y = "Life Expectancy") + geom_point() + labs(title = "Do people in wealthy countries live longer?") + aes(color = continent) + scale_color_manual(values = wes_palette('Cavalcanti1'))
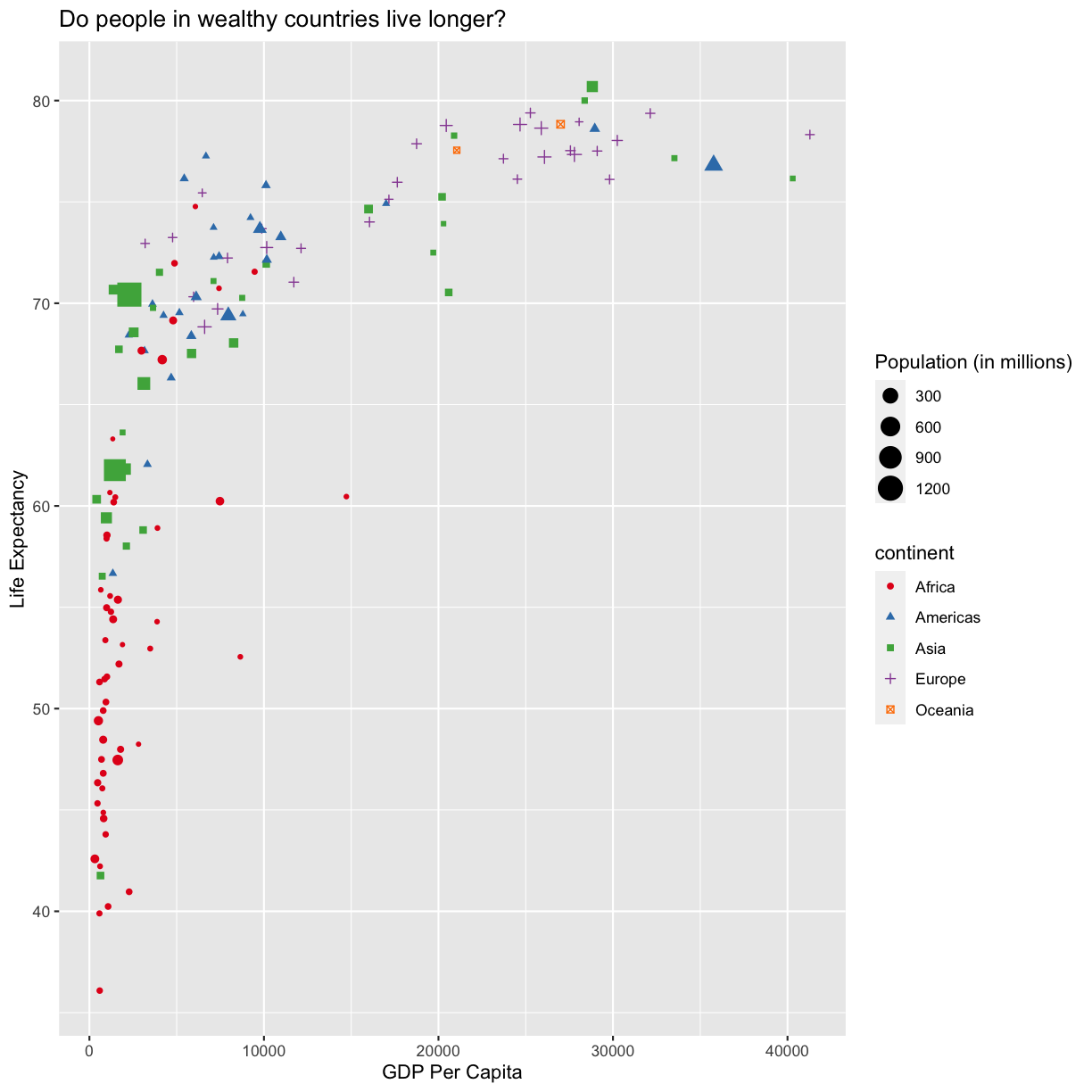
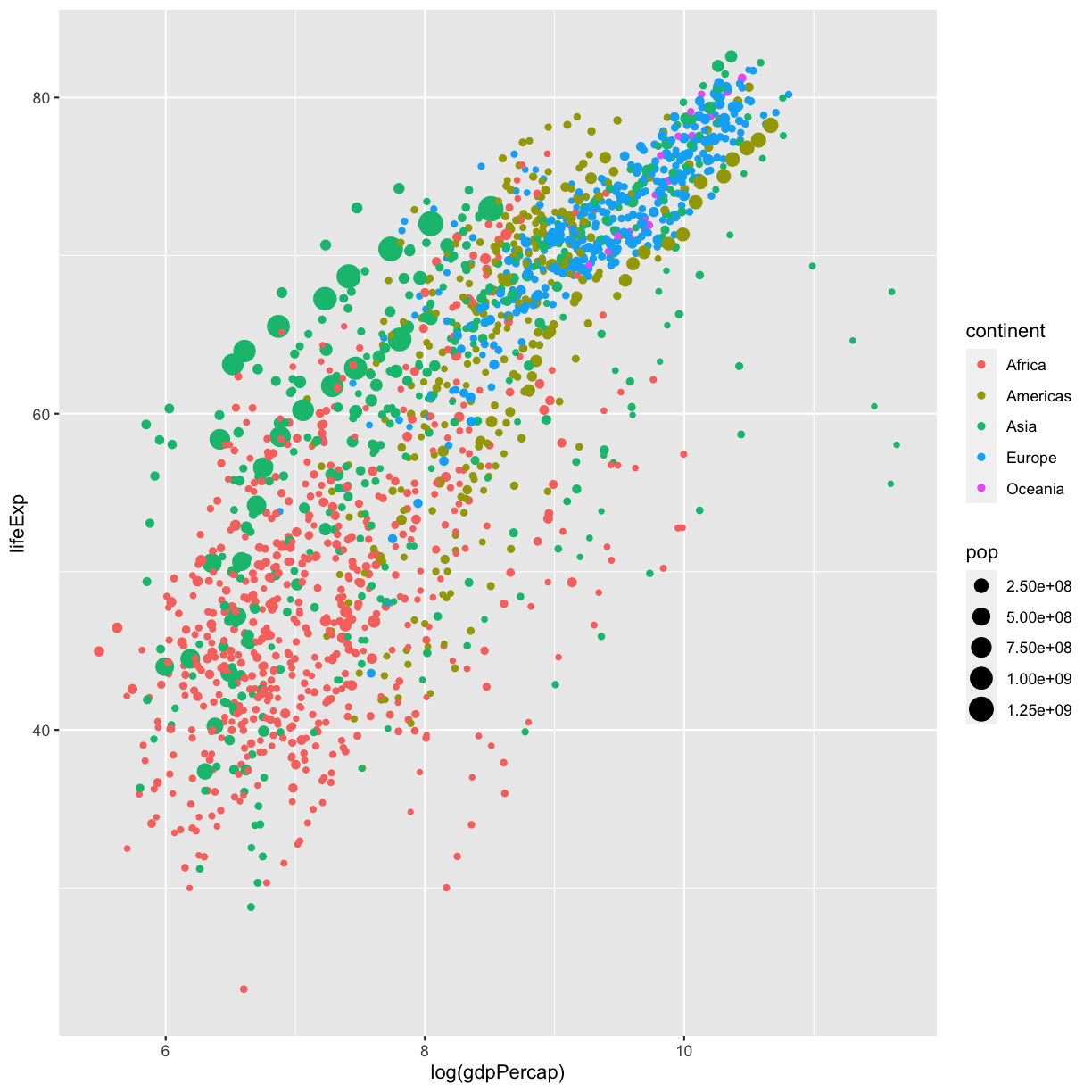
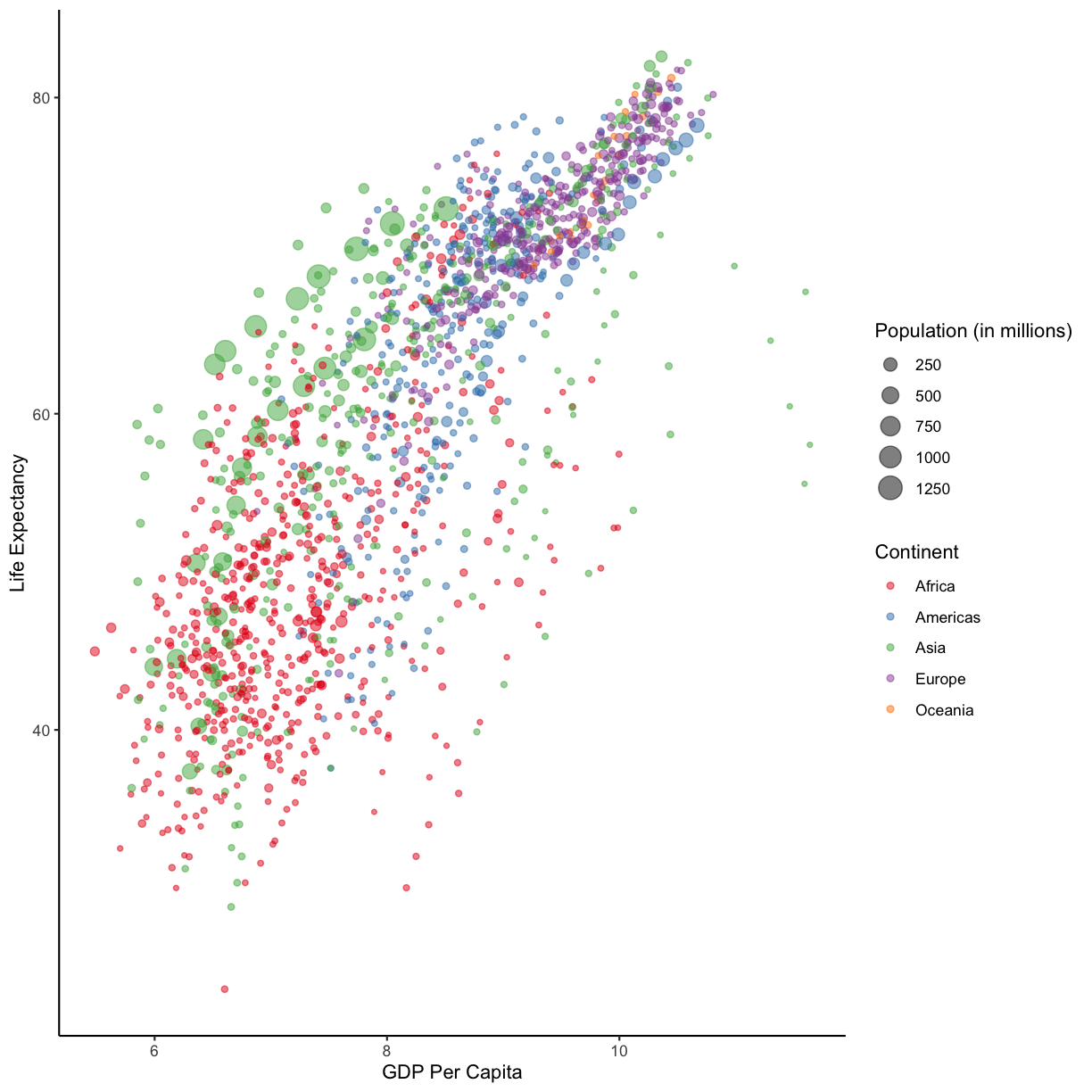
Since we have the data for the population of each country, we might be curious what effect population might have on life expectancy and GDP per capita. Do you think larger countires will have a longer or shorter life expectancy? Let’s find out by mapping the population of each country to the size of our points.
ggplot(data = gapminder_1997) +
aes(x = gdpPercap) +
labs(x = "GDP Per Capita") +
aes(y = lifeExp) +
labs(y = "Life Expectancy") +
geom_point() +
labs(title = "Do people in wealthy countries live longer?") +
aes(color = continent) +
scale_color_brewer(palette = "Set1") +
aes(size = pop)
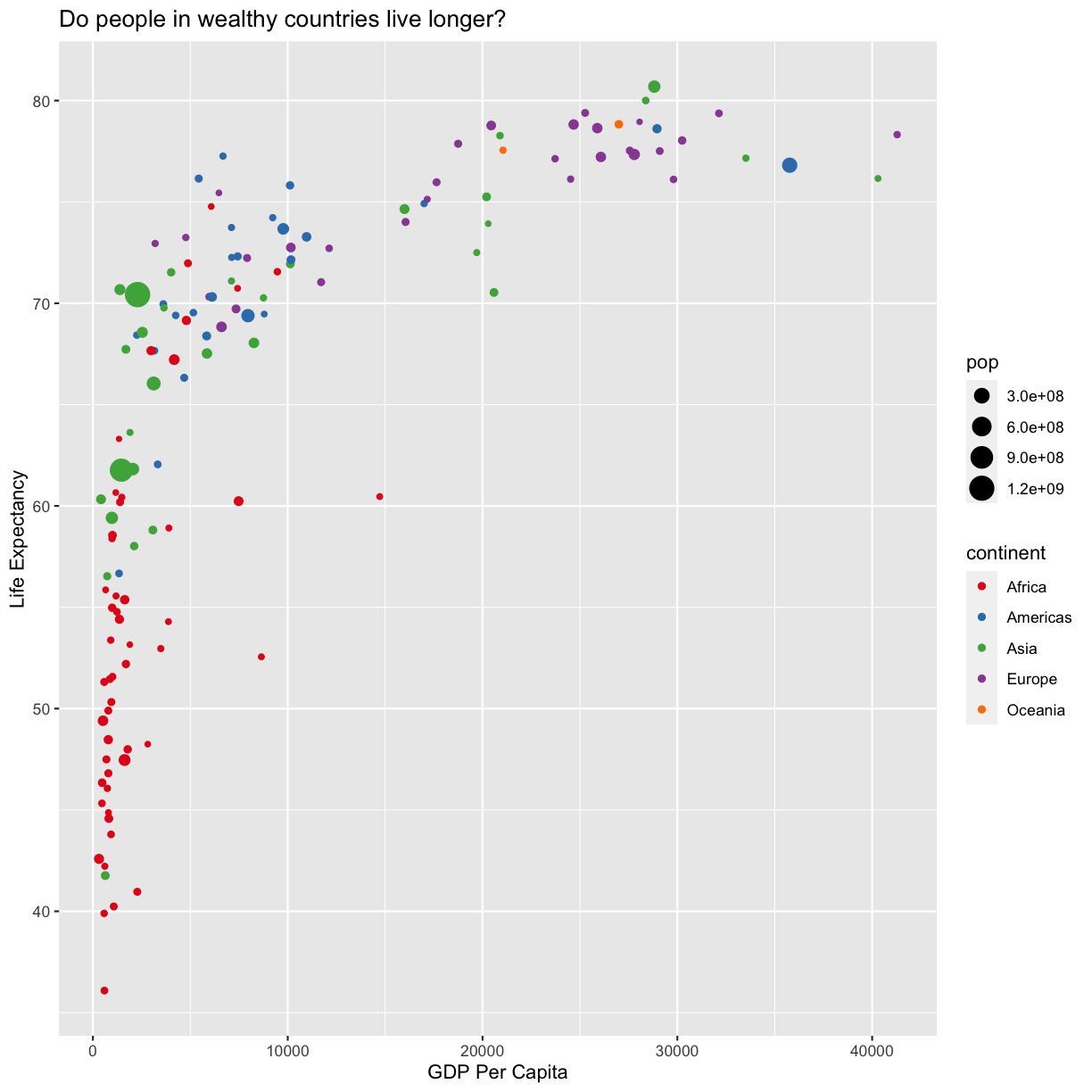
There doesn’t seem to be a very strong association with population size. We can see two very large countries with relatively low GDP per capita (but since the per capita value is already divided by the total population, there is some problems with separating those two values). We got another legend here for size which is nice, but the values look a bit ugly in scientific notation. Let’s divide all the values by 1,000,000 and label our legend “Population (in millions)”
ggplot(data = gapminder_1997) +
aes(x = gdpPercap) +
labs(x = "GDP Per Capita") +
aes(y = lifeExp) +
labs(y = "Life Expectancy") +
geom_point() +
labs(title = "Do people in wealthy countries live longer?") +
aes(color = continent) +
scale_color_brewer(palette = "Set1") +
aes(size = pop/1000000) +
labs(size = "Population (in millions)")
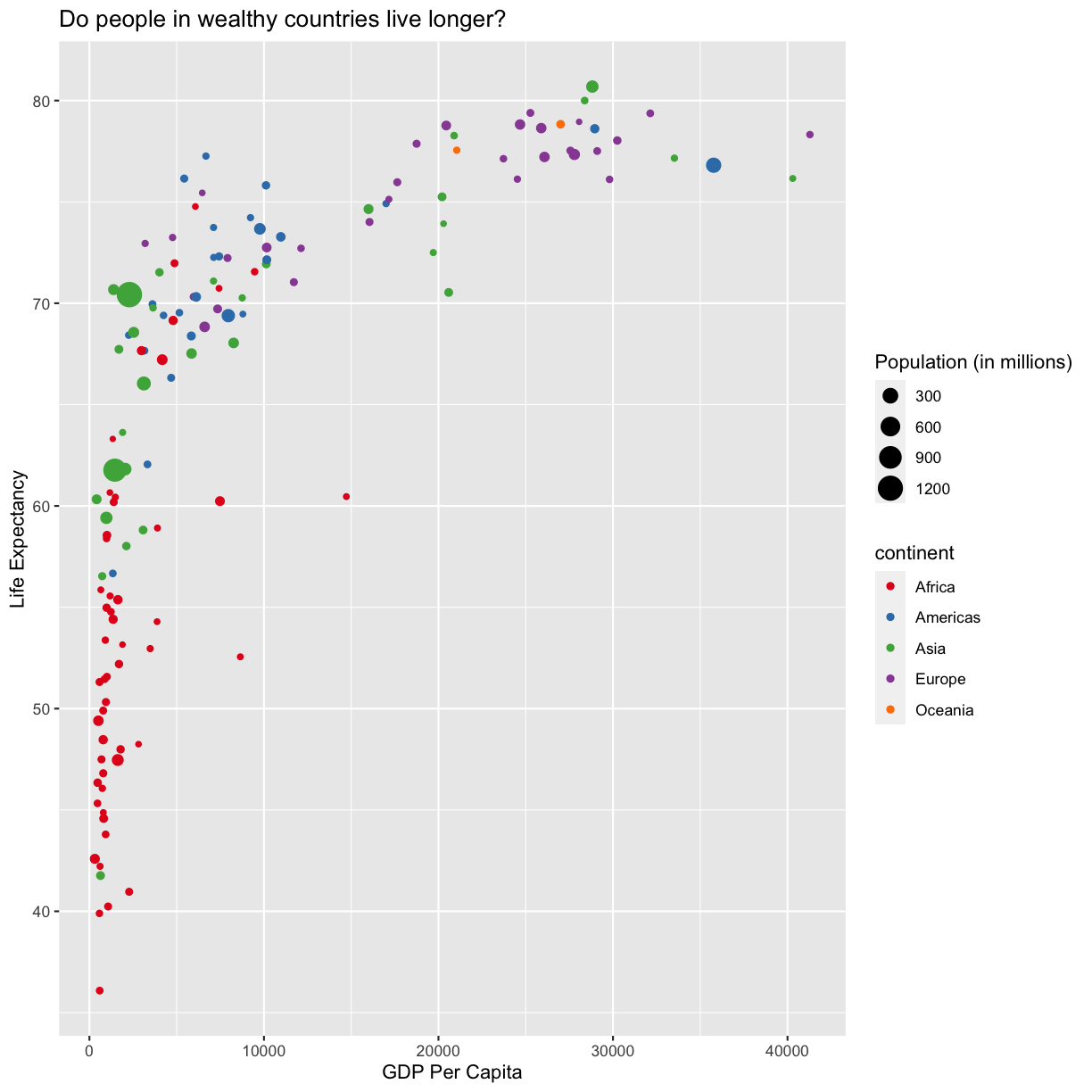
This works because you can treat the columns in the aesthetic mappings just like any other variables and can use functions to transform or change them at plot time rather than having to transform your data first.
Good work! Take a moment to appreciate what a cool plot you made with a few lines of code. In order to fully view its beauty you can click the “Zoom” button in the Plots tab - it will break free from the lower right corner and open the plot in its own window.
Exercise: Changing shapes
Instead of (or in addition to) color, change the shape of the points so each continent has a different shape. (I’m not saying this is a great thing to do - it’s just for practice!) HINT: Is size an aesthetic or a geometry? If you’re stuck, feel free to Google it, or look at the help menu.
Solution
You’ll want to use the
aesaesthetic function to change the shape:ggplot(data = gapminder_1997) + aes(x = gdpPercap) + labs(x = "GDP Per Capita") + aes(y = lifeExp) + labs(y = "Life Expectancy") + geom_point() + labs(title = "Do people in wealthy countries live longer?") + aes(color = continent) + scale_color_brewer(palette = "Set1") + aes(size = pop/1000000) + labs(size = "Population (in millions)") + aes(shape = continent)
For our first plot we added each line of code one at a time so you could see the exact affect it had on the output. But when you start to make a bunch of plots, we can actually combine many of these steps so you don’t have to type as much. For example, you can collect all the aes() statements and all the labs() together. A more condensed version of the exact same plot would look like this:
ggplot(data = gapminder_1997) +
aes(x = gdpPercap, y = lifeExp, color = continent, size = pop/1000000) +
geom_point() +
scale_color_brewer(palette = "Set1") +
labs(x = "GDP Per Capita", y = "Life Expectancy",
title = "Do people in wealthy countries live longer?", size = "Population (in millions)")
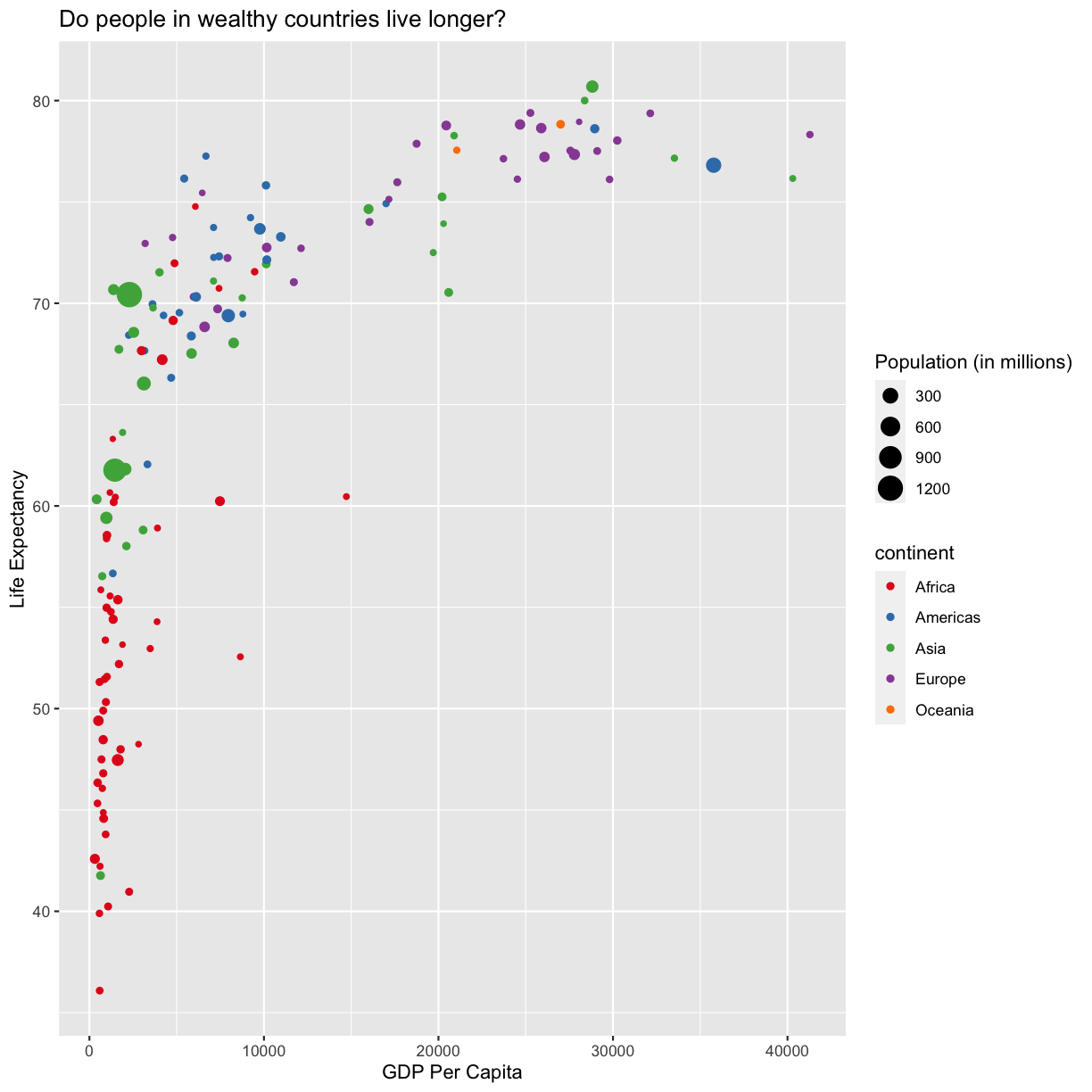
Plotting for data exploration
Many datasets are much more complex than the example we used for the first plot. How can we find meaningful patterns in complex data and create visualizations to convey those patterns?
Importing datasets
In the first plot, we looked at a smaller slice of a large dataset. To gain a better understanding of the kinds of patterns we might observe in our own data, we will now use the full dataset, which is stored in a called “gapminder_data.csv”.
To start, we will read in the data without using the interactive RStudio file navigation.
Rows: 1704 Columns: 6
── Column specification ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
Delimiter: ","
chr (2): country, continent
dbl (4): year, pop, lifeExp, gdpPercap
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
Exercise: Read in your own data
What argument (including file path) should be provided in the below code to read in the full dataset?
gapminder_data <- read_csv()Solution
gapminder_data <- read_csv("gapminder_data.csv")
Let’s take a look at the full dataset. We could use View(), the way we did for the smaller dataset, but if your data is too big, it might take too long to load. Luckily, R offers a way to look at parts of the data to get an idea of what your dataset looks like, without having to examine the whole thing. Here are some commands that allow us to get the dimensions of our data and look at a snapshot of the data. Try them out!
dim(gapminder_data)
head(gapminder_data)
Notice that this dataset has an additional column year compared to the smaller dataset we started with.
Exercise: Predicting
ggplotoutputsNow that we have the full dataset read into our R session, let’s plot the data placing our new
yearvariable on the x axis and life expectancy on the y axis. We’ve provided the code below. Notice that we’ve collapsed the plotting function options and left off some of the labels so there’s not as much code to work with. Before running the code, read through it and see if you can predict what the plot output will look like. Then run the code and check to see if you were right!ggplot(data = gapminder_data) + aes(x=year, y=lifeExp, color=continent) + geom_point()
Hmm, the plot we created in the last challenge isn’t very clear. What’s going on? Since the dataset is more complex, the plotting options we used for the smaller dataset aren’t as useful for interpreting these data. Luckily, we can add additional attributes to our plots that will make patterns more apparent. For example, we can generate a different type of plot - perhaps a line plot - and assign attributes for columns where we might expect to see patterns.
Let’s review the columns and the types of data stored in our dataset to decide how we should group things together. To get an overview of our data object, we can look at the structure of gapminder_data using the str() function.
str(gapminder_data)
(You can also review the structure of your data in the Environment tab by clicking on the blue circle with the arrow in it next to your data object name.)
So, what do we see? The column names are listed after a $ symbol, and then we have a : followed by a text label. These labels correspond to the type of data stored in each column.
What kind of data do we see?
- “int”= Integer (or whole number)
- “num” = Numeric (or non-whole number)
- “chr” = Character (categorical data)
Note In anything before R 4.0, categorical variables used to be read in as factors, which are a special data object that are used to store categorical data and have limited numbers of unique values. The unique values of a factor are tracked via the “levels” of a factor. A factor will always remember all of its levels even if the values don’t actually appear in your data. The factor will also remember the order of the levels and will always print values out in the same order (by default this order is alphabetical).
If your columns are stored as character values but you need factors for plotting, ggplot will convert them to factors for you as needed.
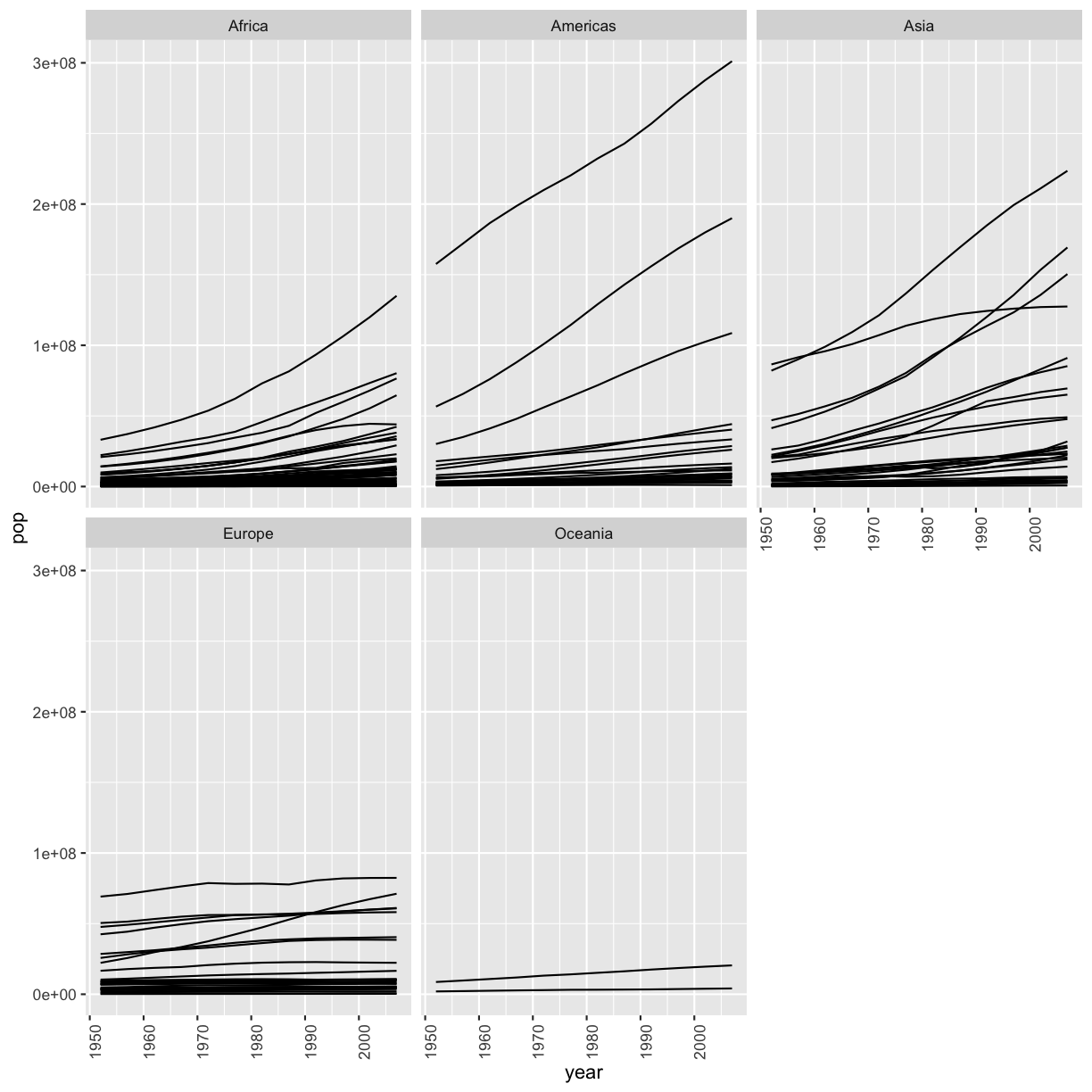
Our plot has a lot of points in columns which makes it hard to see trends over time. A better way to view the data showing changes over time is to use a line plot. Let’s try changing the geom to a line and see what happens.
ggplot(data = gapminder_data) +
aes(x = year, y = lifeExp, color = continent) +
geom_line()
Hmm. This doesn’t look right. By setting the color value, we got a line for each continent, but we really wanted a line for each country. We need to tell ggplot that we want to connect the values for each country value instead. To do this, we need to use the group= aesthetic.
ggplot(data = gapminder_data) +
aes(x = year, y = lifeExp, group = country, color = continent) +
geom_line()
Sometimes plots like this are called “spaghetti plots” because all the lines look like a bunch of wet noodles.
Bonus Exercise: More line plots
Now create your own line plot comparing population and life expectancy! Looking at your plot, can you guess which two countries have experienced massive change in population from 1952-2007?
Solution
ggplot(data = gapminder_data) + aes(x = pop, y = lifeExp, group = country, color = continent) + geom_line()
(China and India are the two Asian countries that have experience massive population growth from 1952-2007.)
Discrete Plots
So far we’ve looked at two plot types (geom_point and geom_line) which work when both the x and y values are numeric. But sometimes you may have one of your values be discrete (a factor or character).
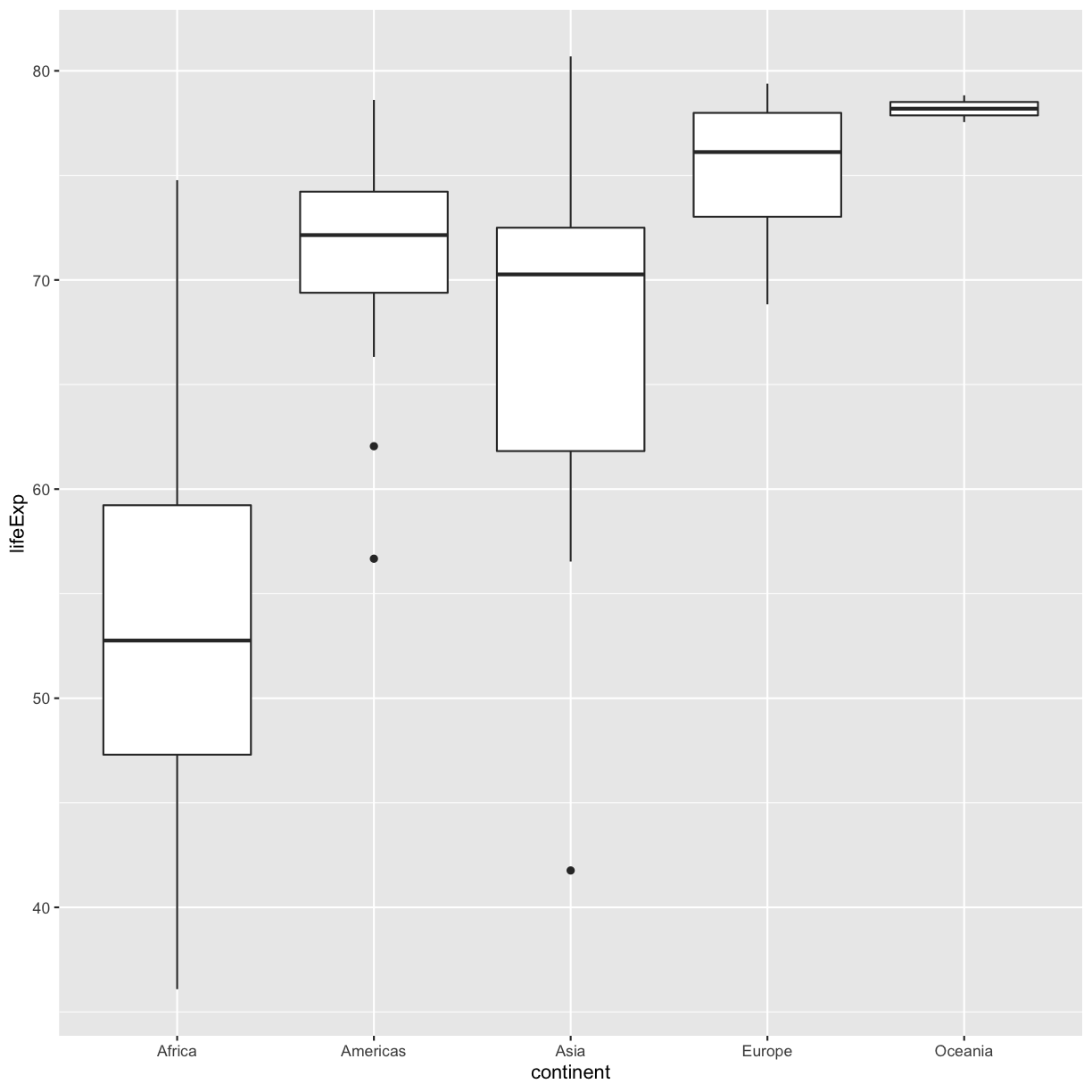
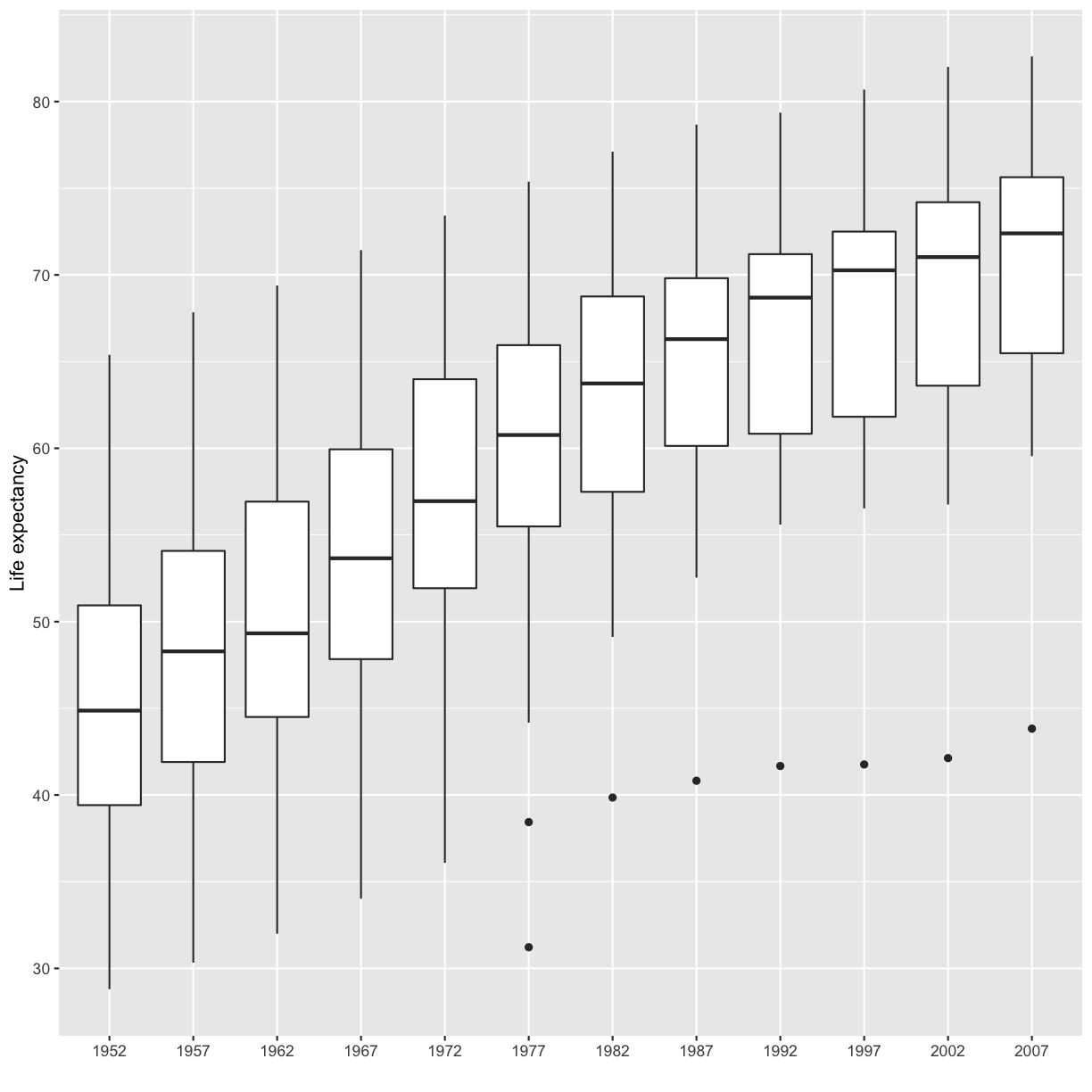
We’ve previously used the discrete values of the continent column to color in our points and lines. But now let’s try moving that variable to the x axis. Let’s say we are curious about comparing the distribution of the life expectancy values for each of the different continents for the gapminder_1997 data. We can do so using a box plot. Try this out yourself in the exercise below!
Exercise: Box plots
Using the
gapminder_1997data, use ggplot to create a box plot with continent on the x axis and life expectancy on the y axis. You can use the examples from earlier in the lesson as a template to remember how to pass ggplot data and map aesthetics and geometries onto the plot. If you’re really stuck, feel free to use the internet as well!Solution
ggplot(data = gapminder_1997) + aes(x = continent, y = lifeExp) + geom_boxplot()
This type of visualization makes it easy to compare the range and spread of values across groups. The “middle” 50% of the data is located inside the box and outliers that are far away from the central mass of the data are drawn as points.
Bonus Exercise:
Take a look a the ggplot cheat sheet. Find all the geoms listed under “Discrete X, Continuous Y”. Try replacing
geom_boxplotwith one of these other functions.Example solution
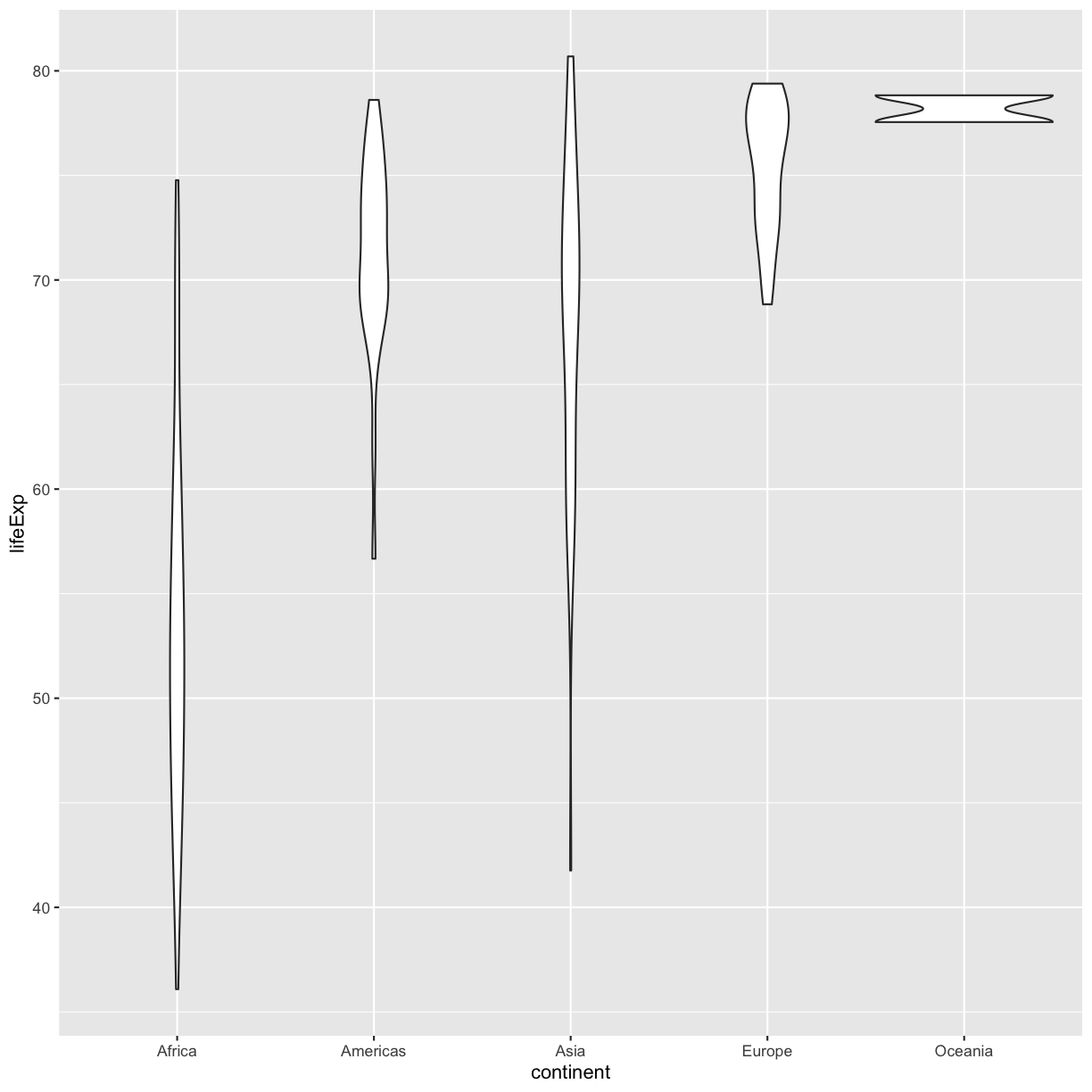
ggplot(data = gapminder_1997) + aes(x = continent, y = lifeExp) + geom_violin()
Layers
So far we’ve only been adding one geom to each plot, but each plot object can actually contain multiple layers and each layer has it’s own geom. Let’s start with a basic violin plot:
ggplot(data = gapminder_1997) +
aes(x = continent, y = lifeExp) +
geom_violin()
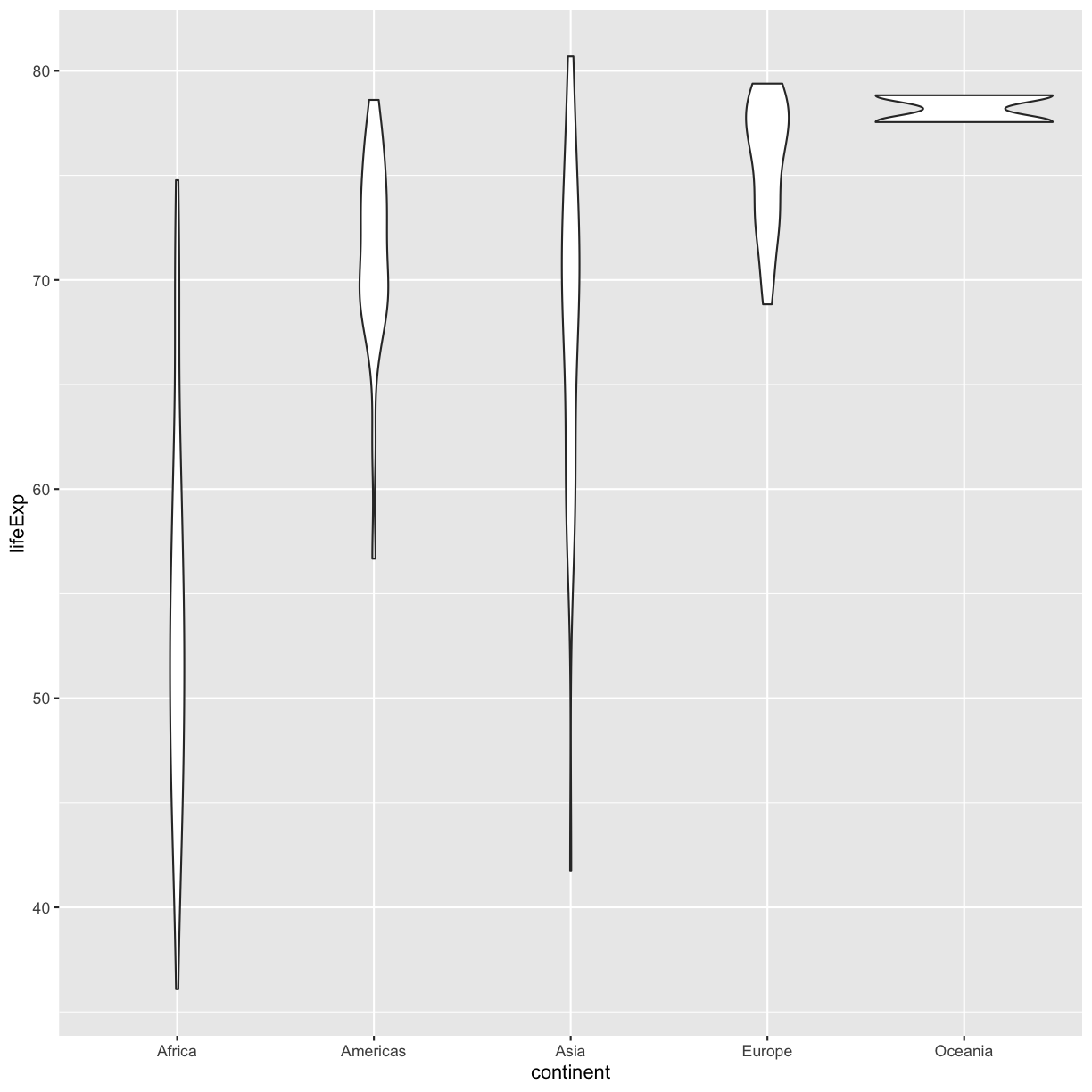
Violin plots are similar to box plots, but they show the range and spread of values with curves rather than boxes (wider curves = more observations) and they do not include outliers. Also note you need a minimum number of points so they can be drawn - because Oceania only has two values, it doesn’t get a curve. We can include the Oceania data by adding a layer of points on top that will show us the “raw” data:
ggplot(data = gapminder_1997) +
aes(x = continent, y = lifeExp) +
geom_violin() +
geom_point()
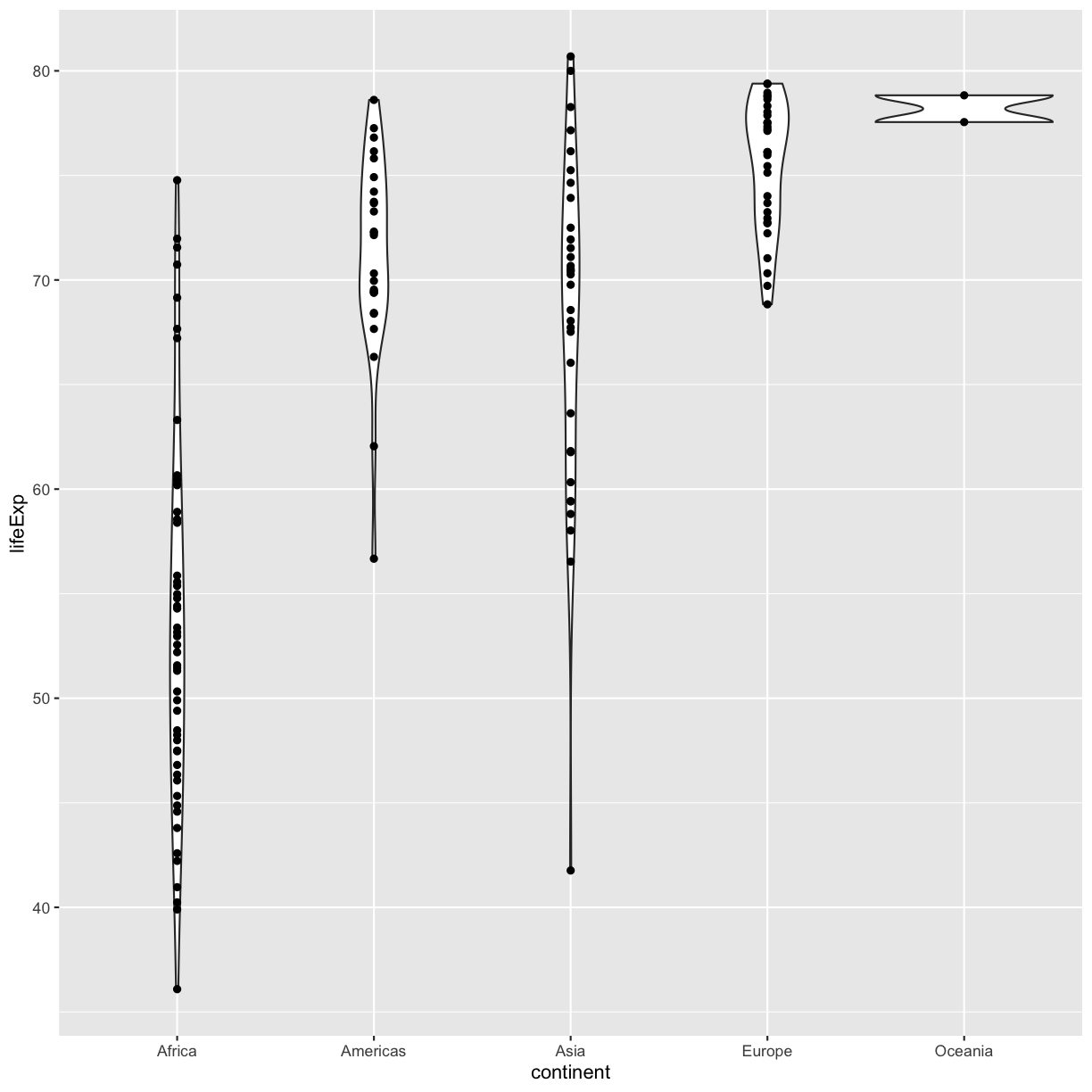
OK, we’ve drawn the points but most of them stack up on top of each other. One way to make it easier to see all the data is to “jitter” the points, or move them around randomly so they don’t stack up on top of each other. To do this, we use geom_jitter rather than geom_point
ggplot(data = gapminder_1997) +
aes(x = continent, y = lifeExp) +
geom_violin() +
geom_jitter()
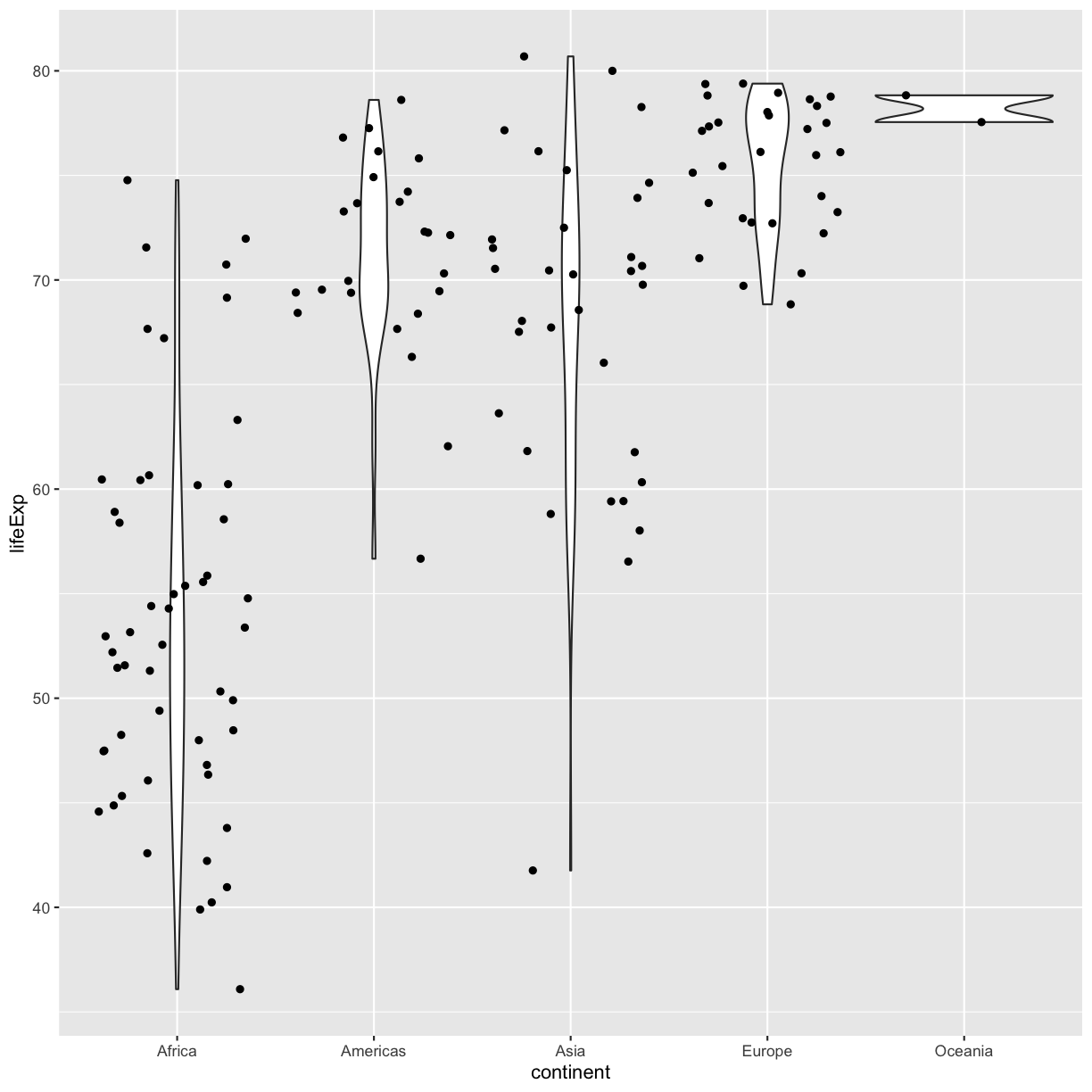
Be aware that these movements are random so your plot will look a bit different each time you run it!
Now let’s try switching the order of geom_violin and geom_jitter. What happens? Why?
ggplot(data = gapminder_1997) +
aes(x = continent, y = lifeExp) +
geom_jitter() +
geom_violin()
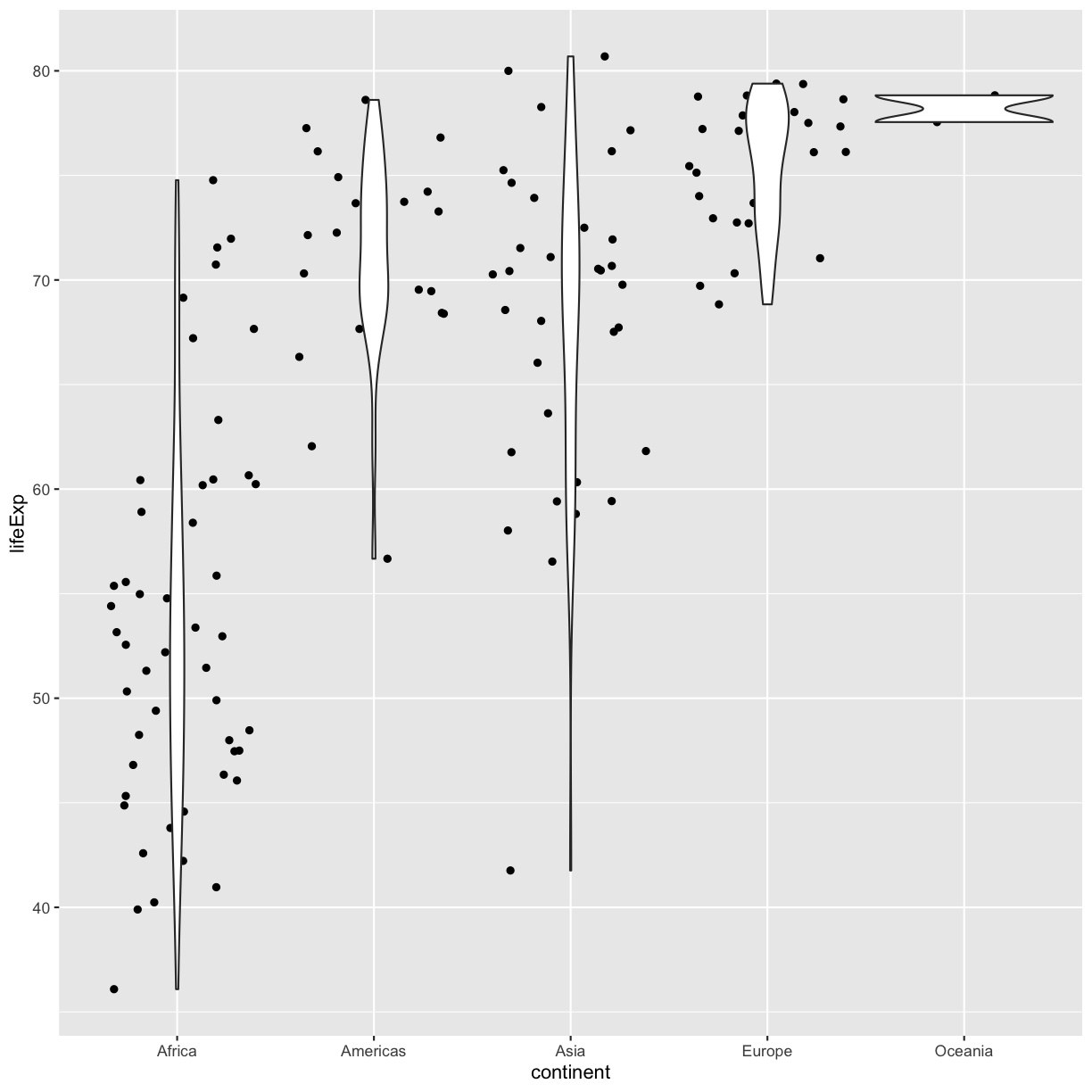
Since we plot the geom_jitter layer first, the violin plot layer is placed on top of the geom_jitter layer, so we cannot see most of the points.
Note that each layer can have it’s own set of aesthetic mappings. So far we’ve been using aes() outside of the other functions. When we do this, we are setting the “default” aesthetic mappings for the plot. We could do the same thing by passing the values to the ggplot() function call as is sometimes more common:
ggplot(data = gapminder_1997, mapping = aes(x = continent, y = lifeExp)) +
geom_violin() +
geom_jitter()
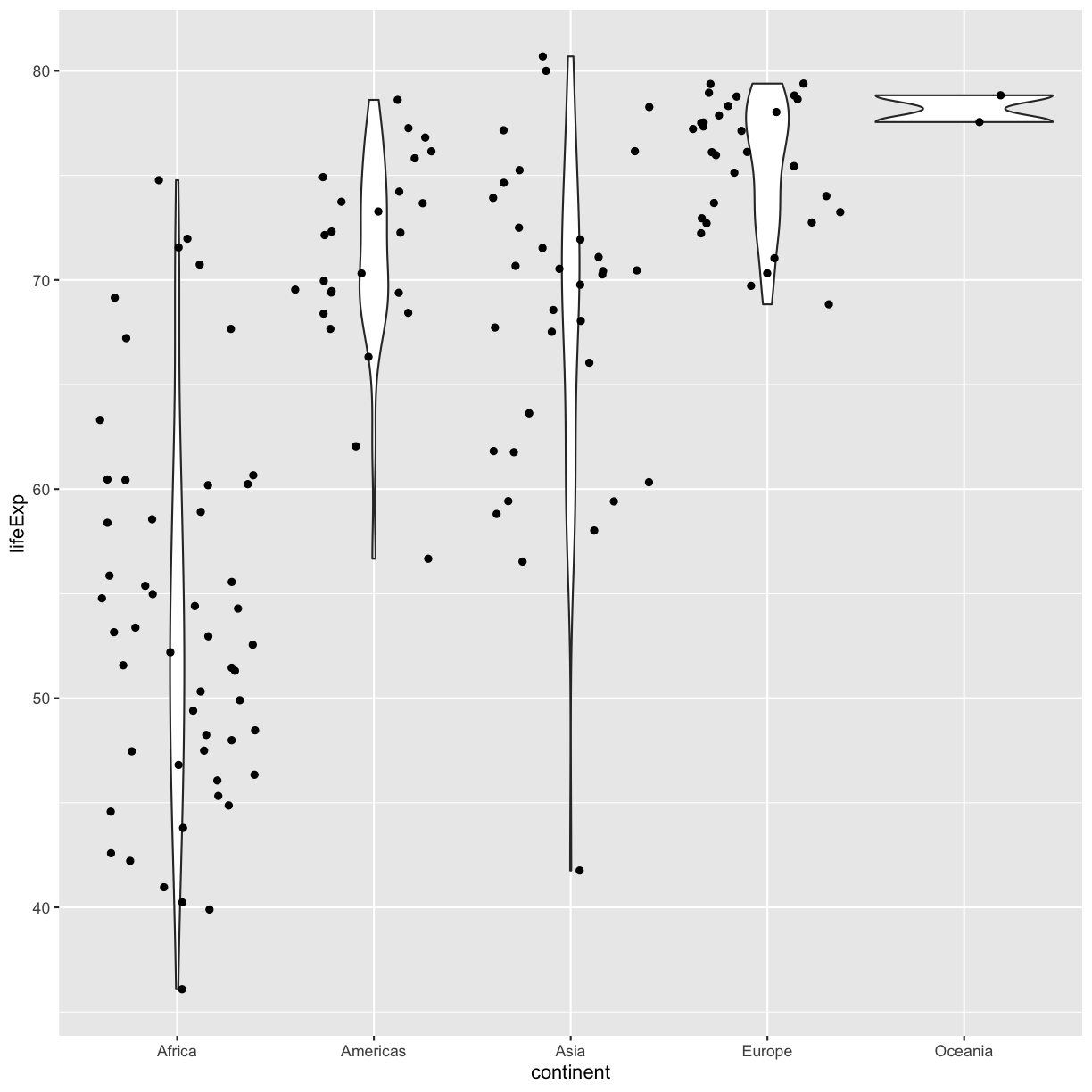
However, we can also use aesthetic values for only one layer of our plot. To do that, you an place an additional aes() inside of that layer. For example, what if we want to change the size for the points so they are scaled by population, but we don’t want to change the violin plot? We can do:
ggplot(data = gapminder_1997) +
aes(x = continent, y = lifeExp) +
geom_violin() +
geom_jitter(aes(size = pop))
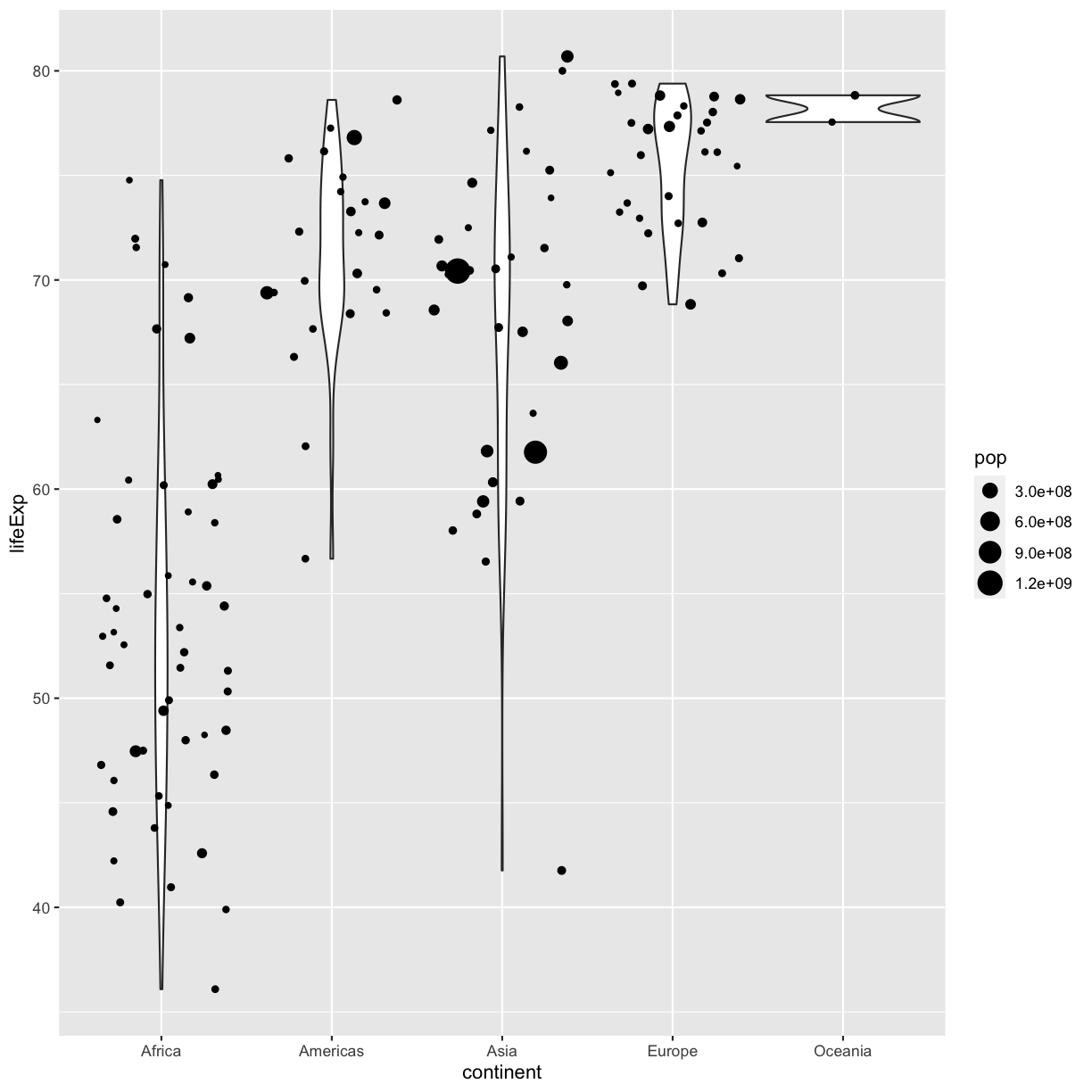
Both geom_violin and geom_jitter will inherit the default values of aes(continent, lifeExp) but only geom_jitter will also use aes(size = pop).
Functions within functions
In the two examples above, we placed the
aes()function inside another function - see how in the line of codegeom_jitter(aes(size = pop)),aes()is nested insidegeom_jitter()? When this happens, R evaluates the inner function first, then passes the output of that function as an argument to the outer function.Take a look at this simpler example. Suppose we have:
sum(2, max(6,8))First R calculates the maximum of the numbers 6 and 8 and returns the value 8. It passes the output 8 into the sum function and evaluates:
sum(2, 8)[1] 10
Color vs. Fill
Let’s say we want to spice up our plot a bit by adding some color. Maybe we want our violin color to a fancy color like “darkolivegreen.” We can do this by explicitly setting the color aesthetic inside the geom_violin function. Note that because we are assigning a color directly and not using any values from our data to do so, we do not need to use the aes() mapping function. Let’s try it out:
ggplot(data = gapminder_1997) +
aes(x = continent, y = lifeExp) +
geom_violin(color="darkolivegreen")
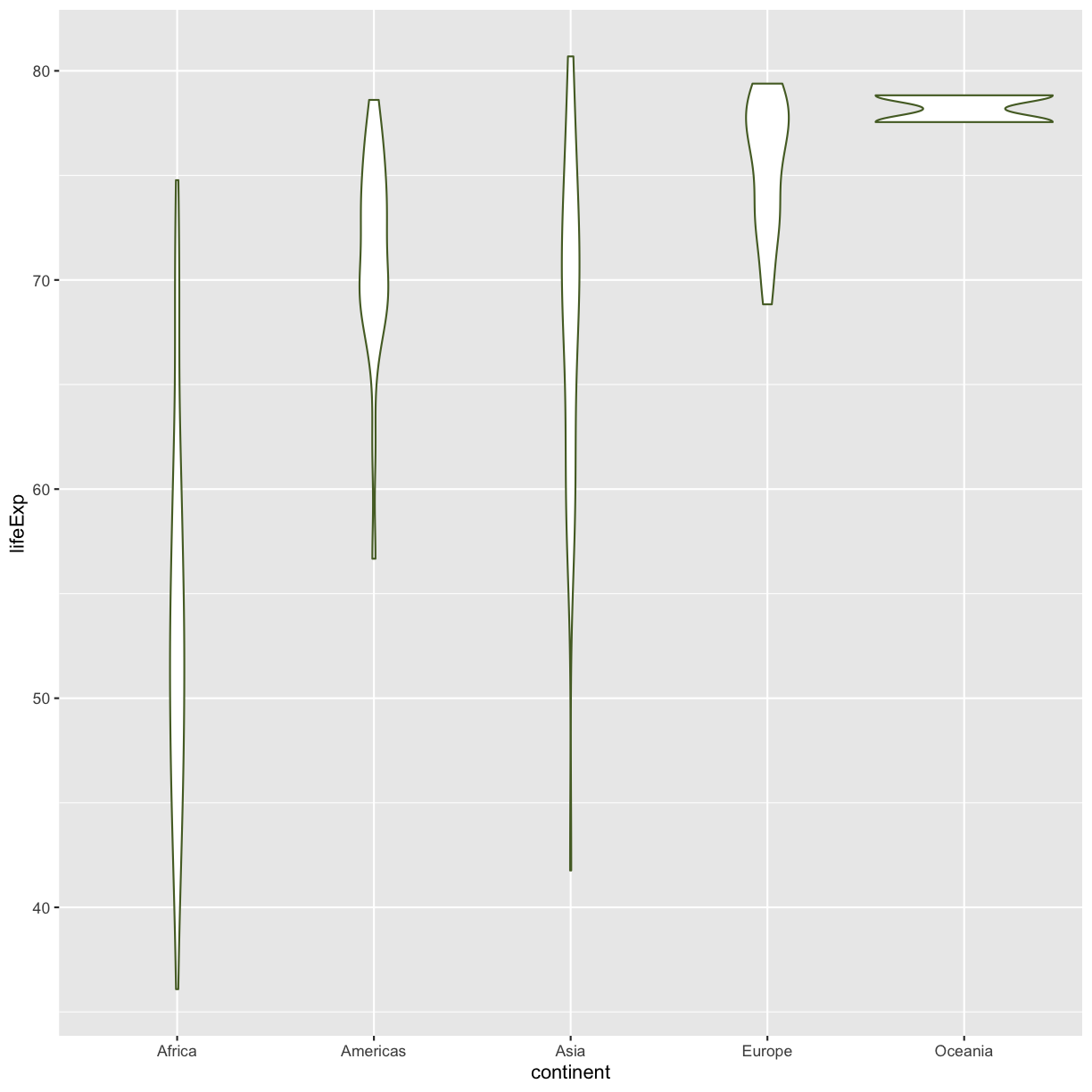 Well, that didn’t get all that colorful. That’s because objects like these violins have two different parts that have a color: the shape outline, and the inner part of the shape. For geoms that have an inner part, you change the fill color with
Well, that didn’t get all that colorful. That’s because objects like these violins have two different parts that have a color: the shape outline, and the inner part of the shape. For geoms that have an inner part, you change the fill color with fill= rather than color=, so let’s try that instead
ggplot(data = gapminder_1997) +
aes(x = continent, y = lifeExp) +
geom_violin(fill="darkolivegreen")
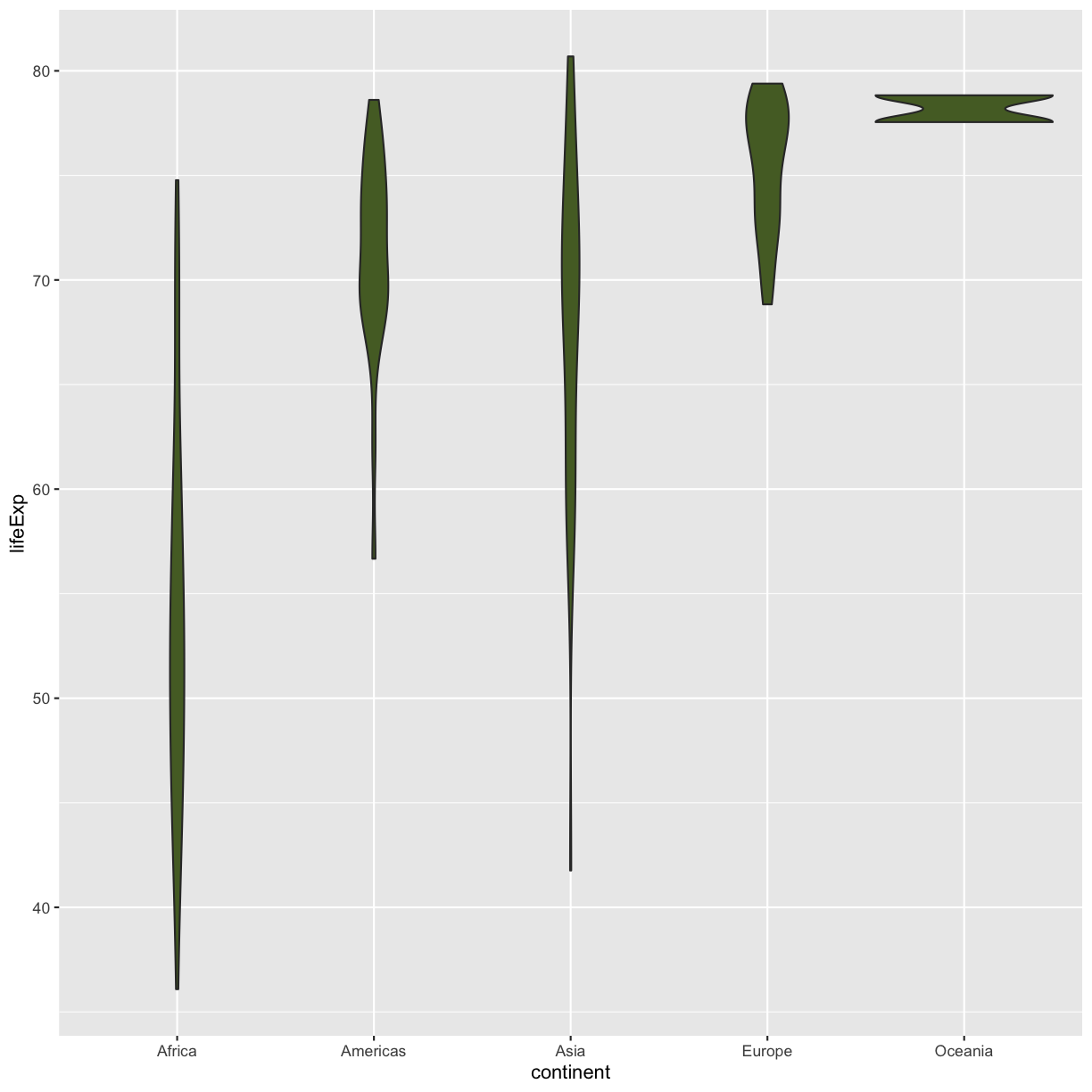
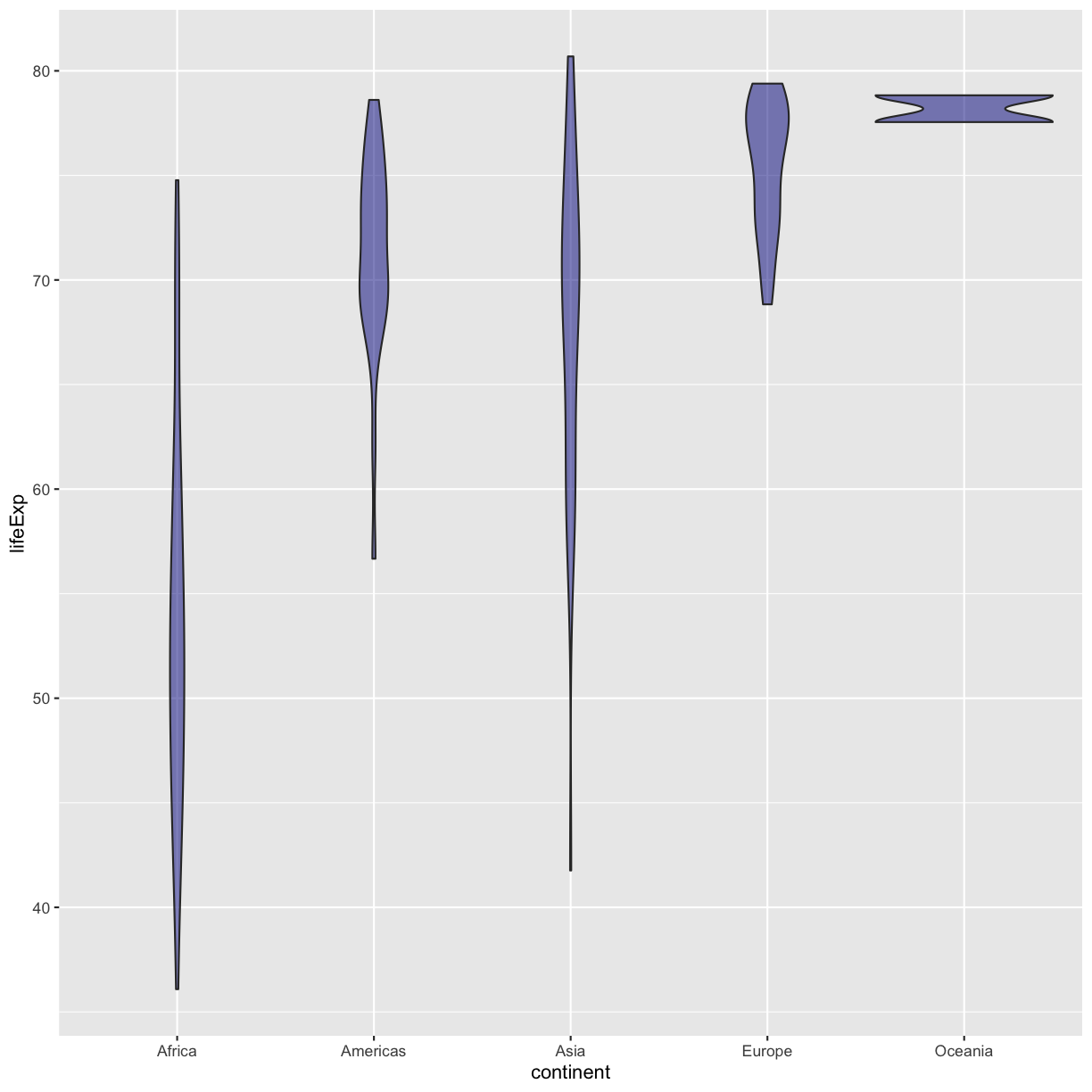
That’s some plot now isn’t it! Compare this to what you see when you map the fill property to your data rather than setting a specific value.
ggplot(data = gapminder_1997) +
aes(x = continent, y = lifeExp) +
geom_violin(aes(fill=continent))
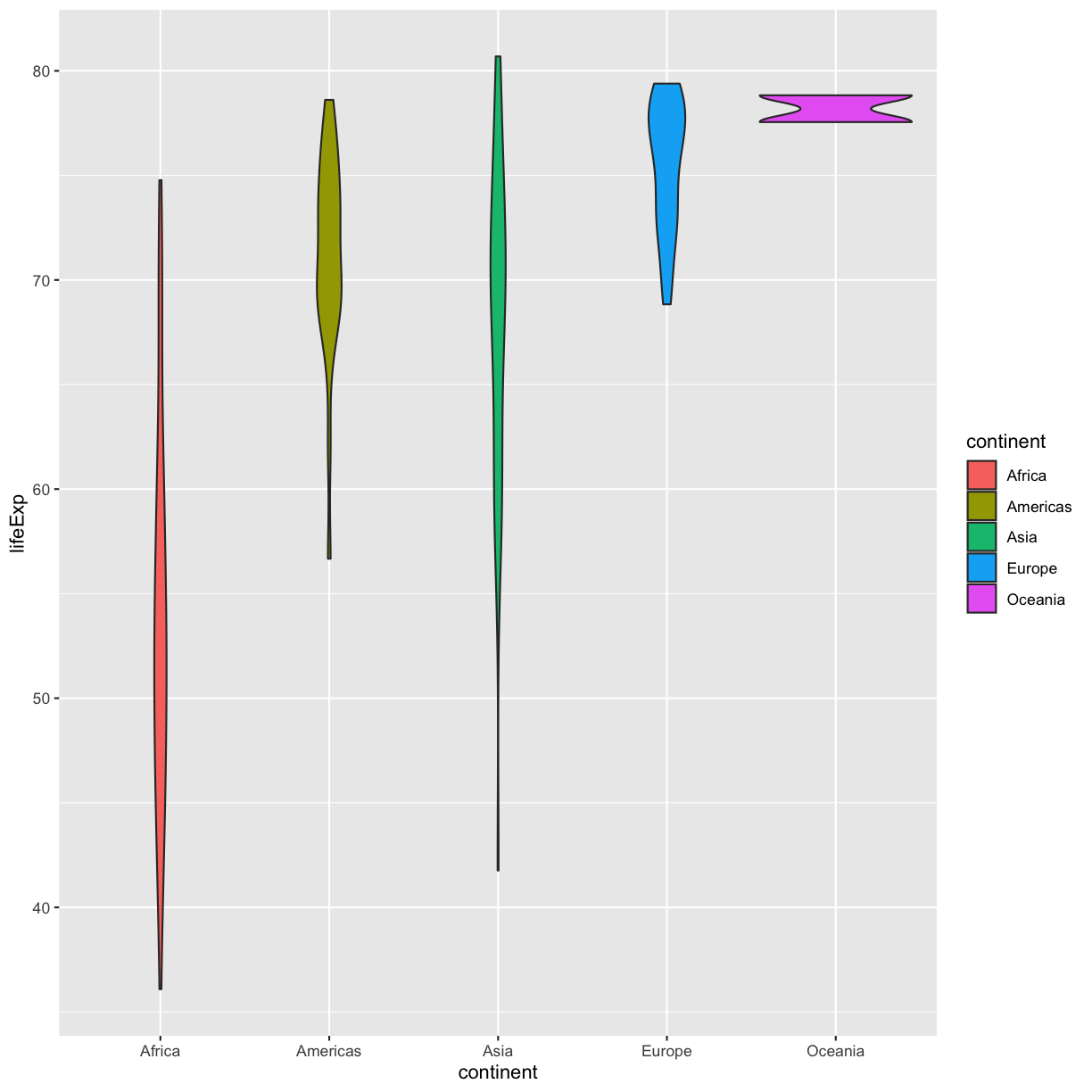
So “darkolivegreen” maybe wasn’t the prettiest color. R knows lots of color names. You can see the full list if you run colors() in the console. Since there are so many, you can randomly choose 10 if you run sample(colors(), size = 10).
Exercise: choosing a color
Use
sample(colors(), size = 10)a few times until you get an interesting sounding color name and swap that out for “darkolivegreen” in the violin plot example.
Bonus Exercise: Transparency
Another aesthetic that can be changed is how transparent our colors/fills are. The
alphaparameter decides how transparent to make the colors. By default,alpha = 1, and our colors are completely opaque. Decreasingalphaincreases the transparency of our colors/fills. Try changing the transparency of our violin plot. (Hint: Should alpha be inside or outsideaes()?)Solution
ggplot(data = gapminder_1997) + aes(x = continent, y = lifeExp) + geom_violin(fill="darkblue", alpha = 0.5)
Changing colors
What happens if you run:
ggplot(data = gapminder_1997) + aes(x = continent, y = lifeExp) + geom_violin(aes(fill = "springgreen"))
Why doesn’t this work? How can you fix it? Where does that color come from?
Solution
In this example, you placed the fill inside the
aes()function. Because you are using an aesthetic mapping, the “scale” for the fill will assign colors to values - in this case, you only have one value: the word “springgreen.” Instead, trygeom_violin(fill = "springgreen").
Univariate Plots
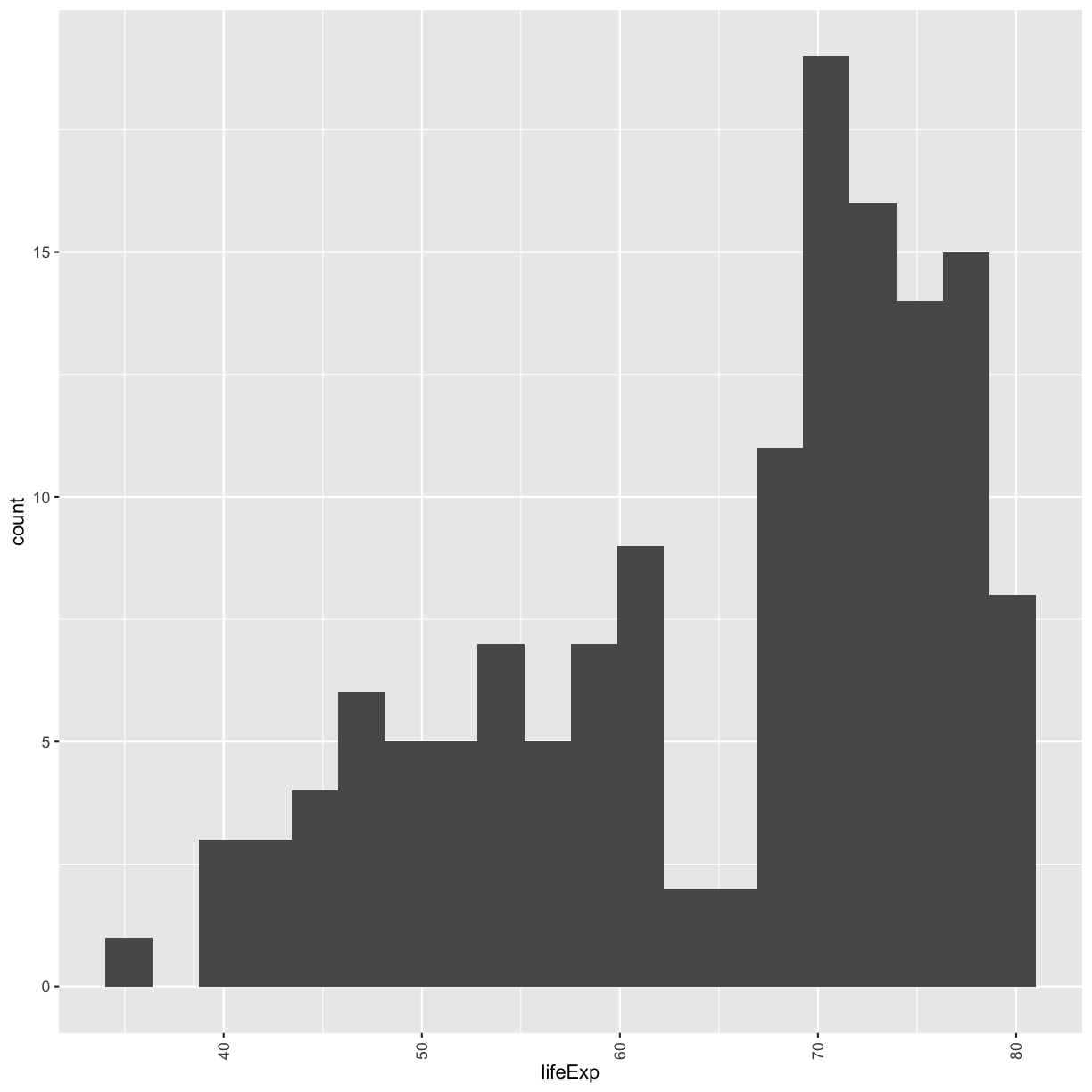
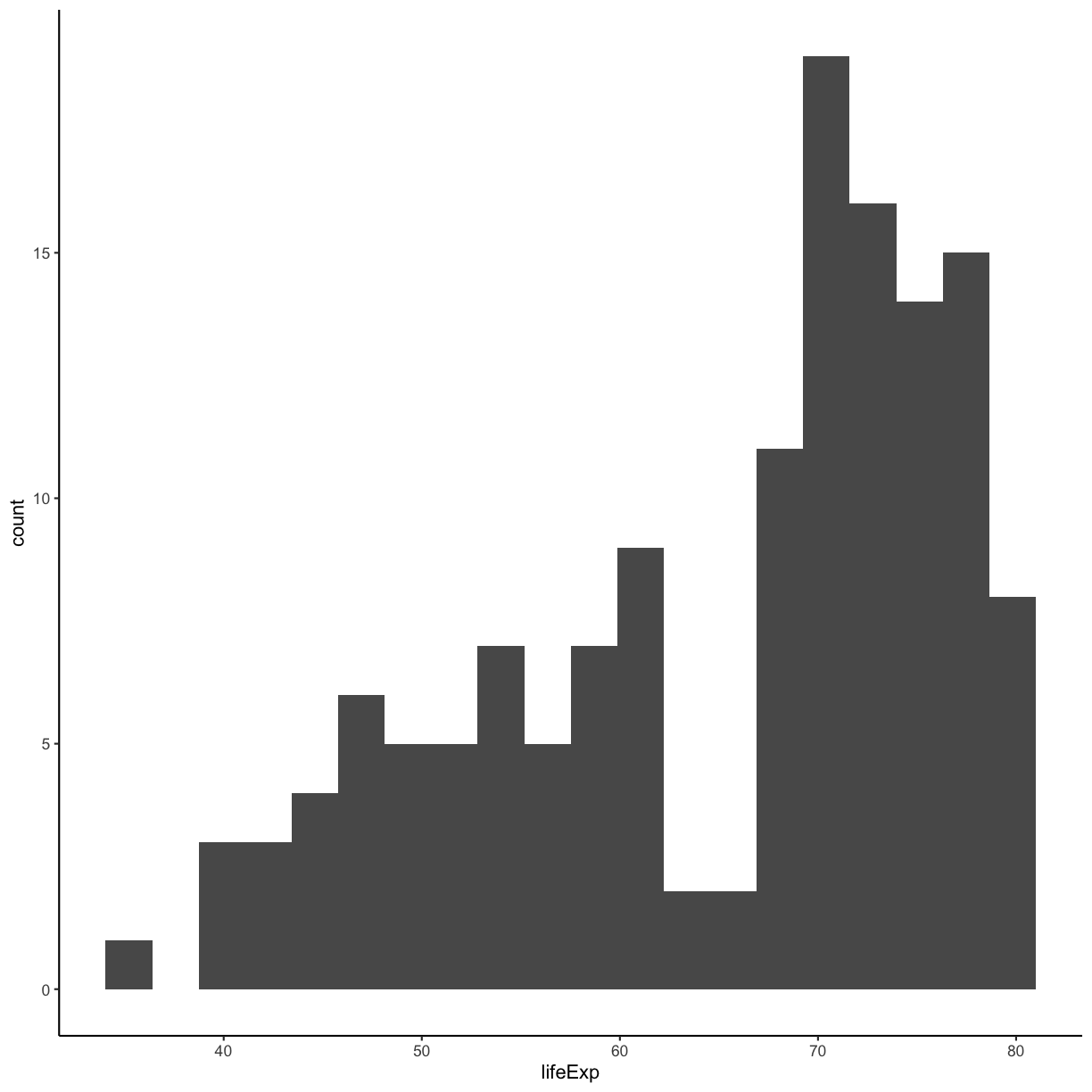
We jumped right into make plots with multiple columns. But what if we wanted to take a look at just one column? In that case, we only need to specify a mapping for x and choose an appropriate geom. Let’s start with a histogram to see the range and spread of the life expectancy values
ggplot(gapminder_1997) +
aes(x = lifeExp) +
geom_histogram()
`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
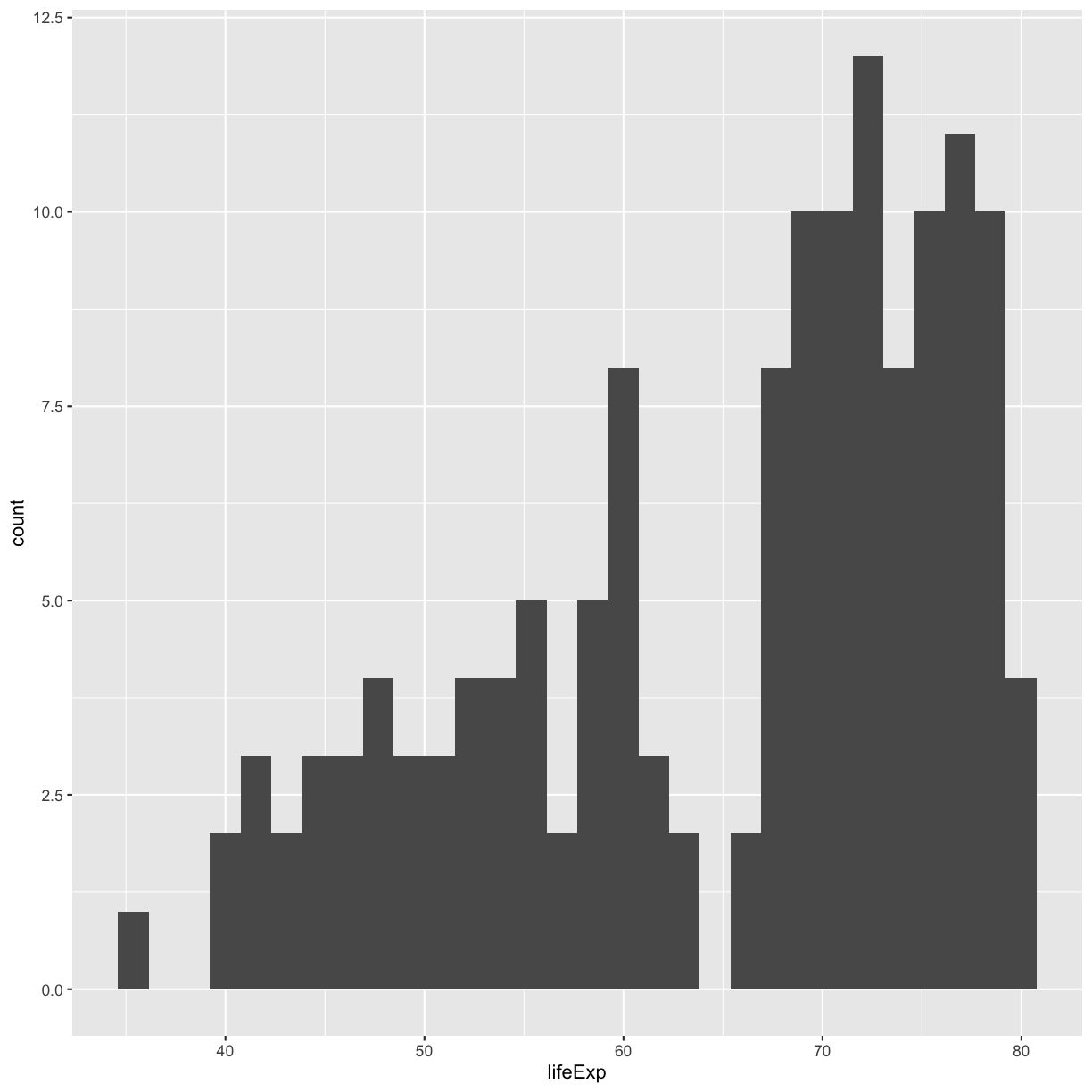
You should not only see the plot in the plot window, but also a message telling you to choose a better bin value. Histograms can look very different depending on the number of bars you decide to draw. The default is 30. Let’s try setting a different value by explicitly passing a bin= argument to the geom_histogram later.
ggplot(gapminder_1997) +
aes(x = lifeExp) +
geom_histogram(bins=20)
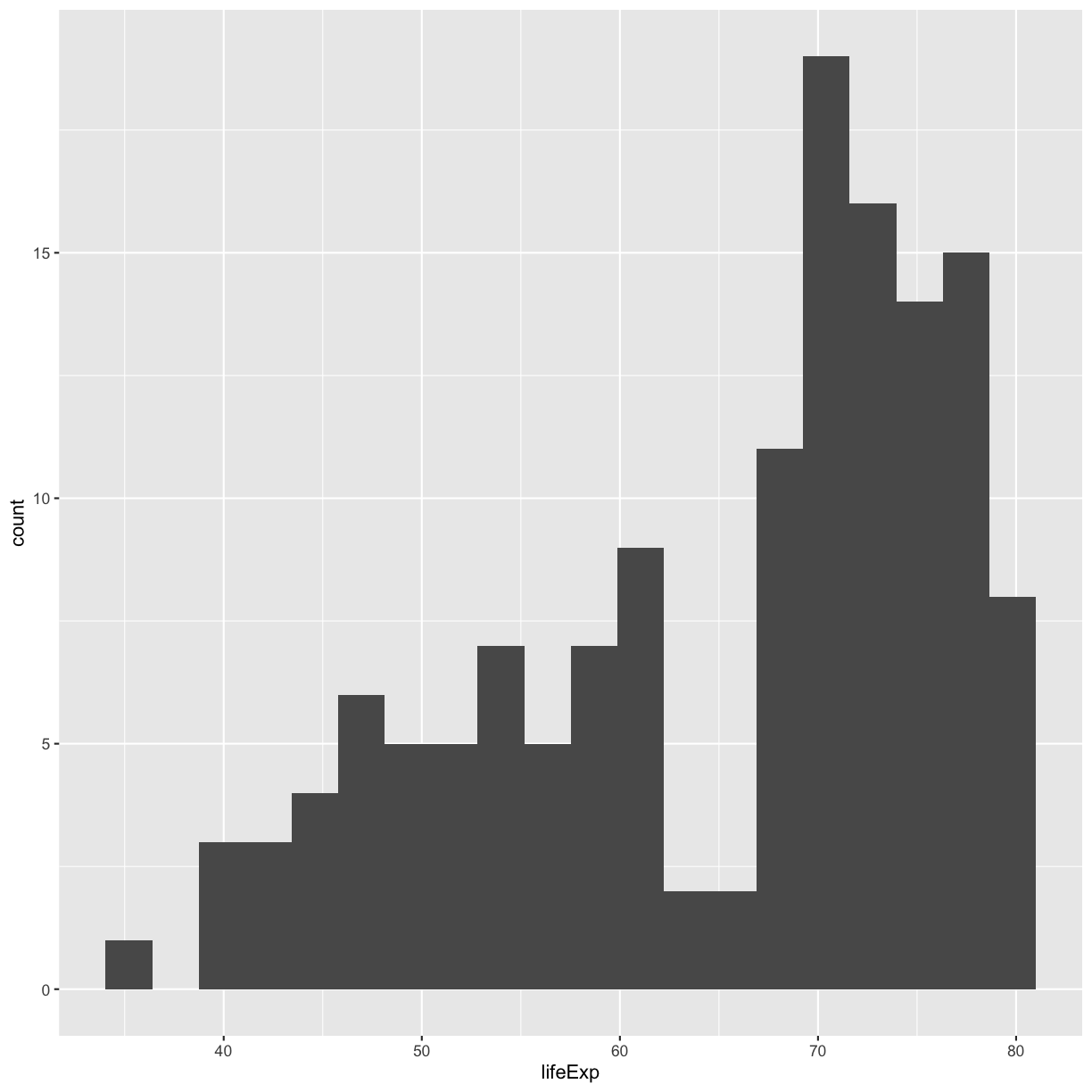
Try different values like 5 or 50 to see how the plot changes.
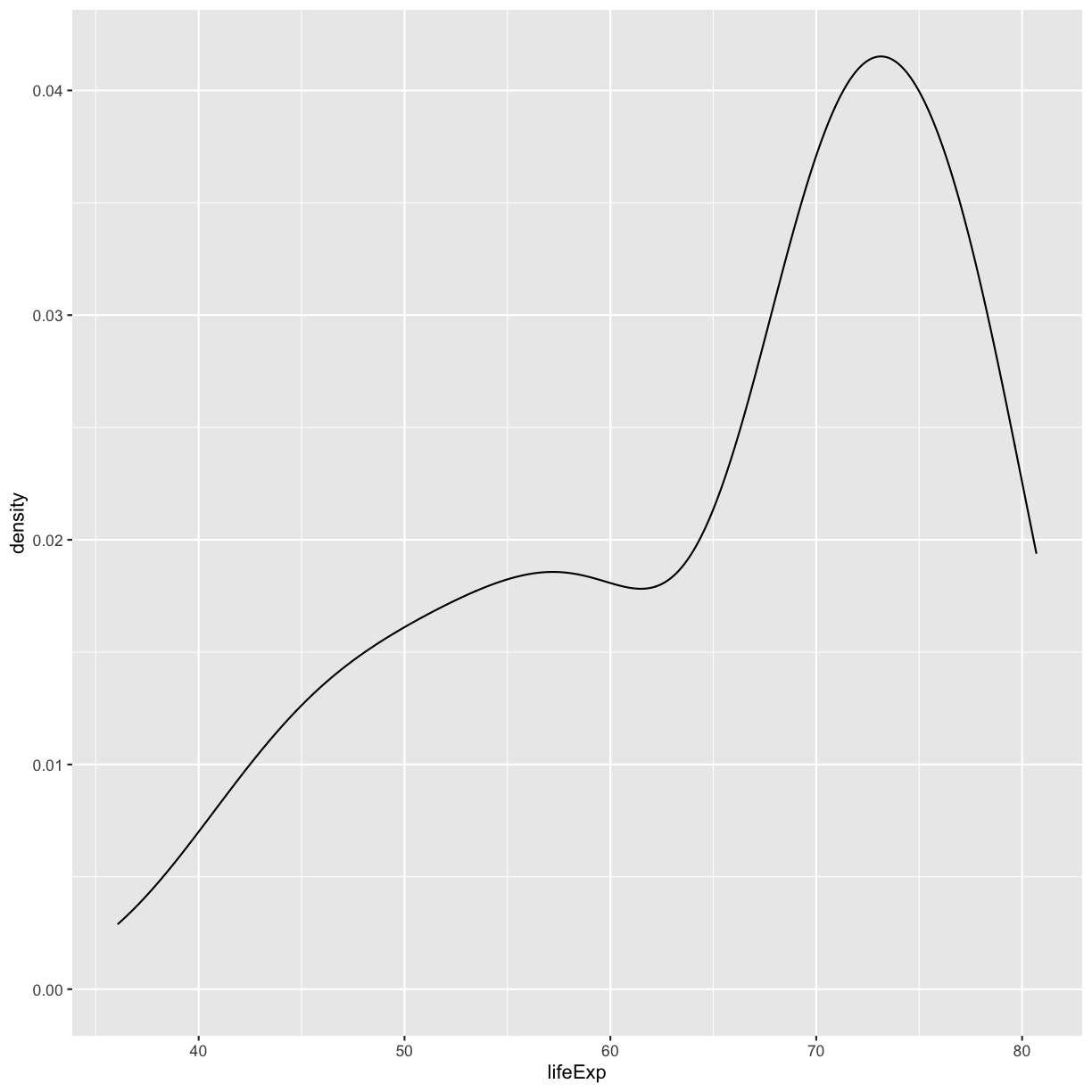
Bonus Exercise: One variable plots
Rather than a histogram, choose one of the other geometries listed under “One Variable” plots on the ggplot cheat sheet. Note that we used
lifeExphere which has continuous values. If you want to try the discrete options, try mappingcontinentto x instead.Example solution
ggplot(gapminder_1997) + aes(x = lifeExp) + geom_density()
Plot Themes
Our plots are looking pretty nice, but what’s with that grey background? While you can change various elements of a ggplot object manually (background color, grid lines, etc.) the ggplot package also has a bunch of nice built-in themes to change the look of your graph. For example, let’s try adding theme_classic() to our histogram:
ggplot(gapminder_1997) +
aes(x = lifeExp) +
geom_histogram(bins = 20) +
theme_classic()
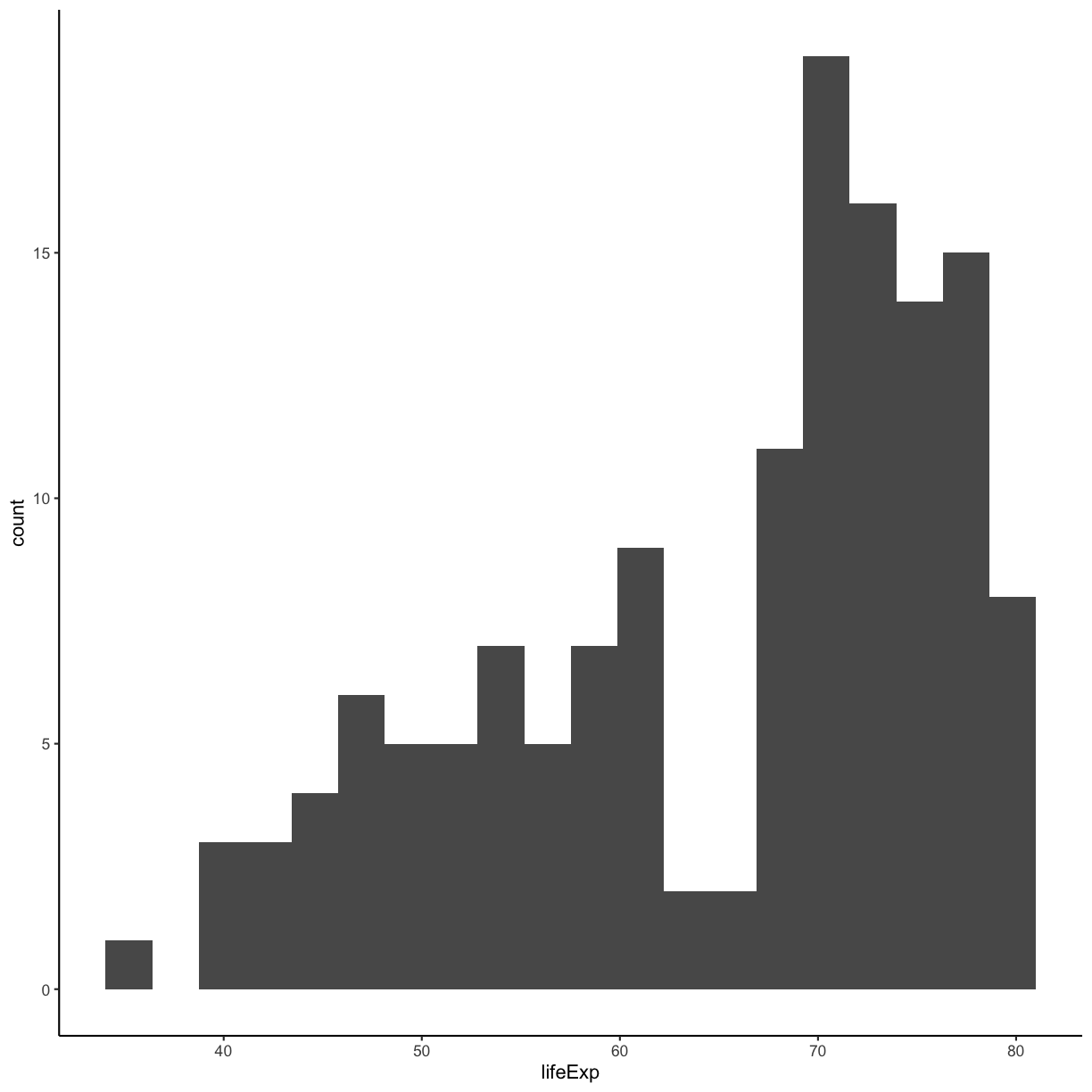
Try out a few other themes, to see which you like: theme_bw(), theme_linedraw(), theme_minimal().
Exercise: Rotating x axis labels
Often, you’ll want to change something about the theme that you don’t know how to do off the top of your head. When this happens, you can do an Internet search to help find what you’re looking for. To practice this, search the Internet to figure out how to rotate the x axis labels 90 degrees. Then try it out using the histogram plot we made above.
Solution
ggplot(gapminder_1997) + aes(x = lifeExp) + geom_histogram(bins = 20) + theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1))
Facets
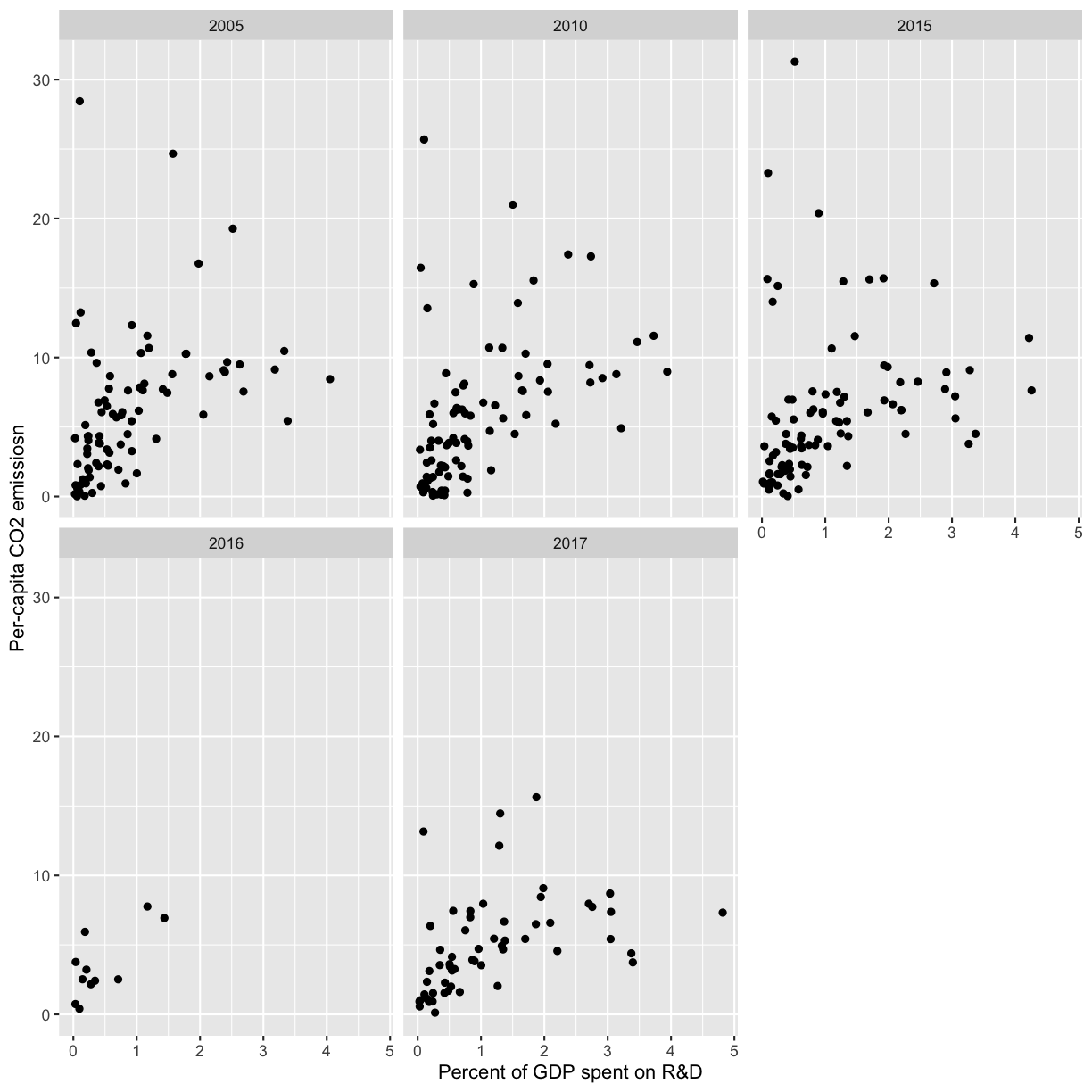
If you have a lot of different columns to try to plot or have distinguishable subgroups in your data, a powerful plotting technique called faceting might come in handy. When you facet your plot, you basically make a bunch of smaller plots and combine them together into a single image. Luckily, ggplot makes this very easy. Let’s start with a simplified version of our first plot
ggplot(gapminder_1997) +
aes(x = gdpPercap, y = lifeExp) +
geom_point()
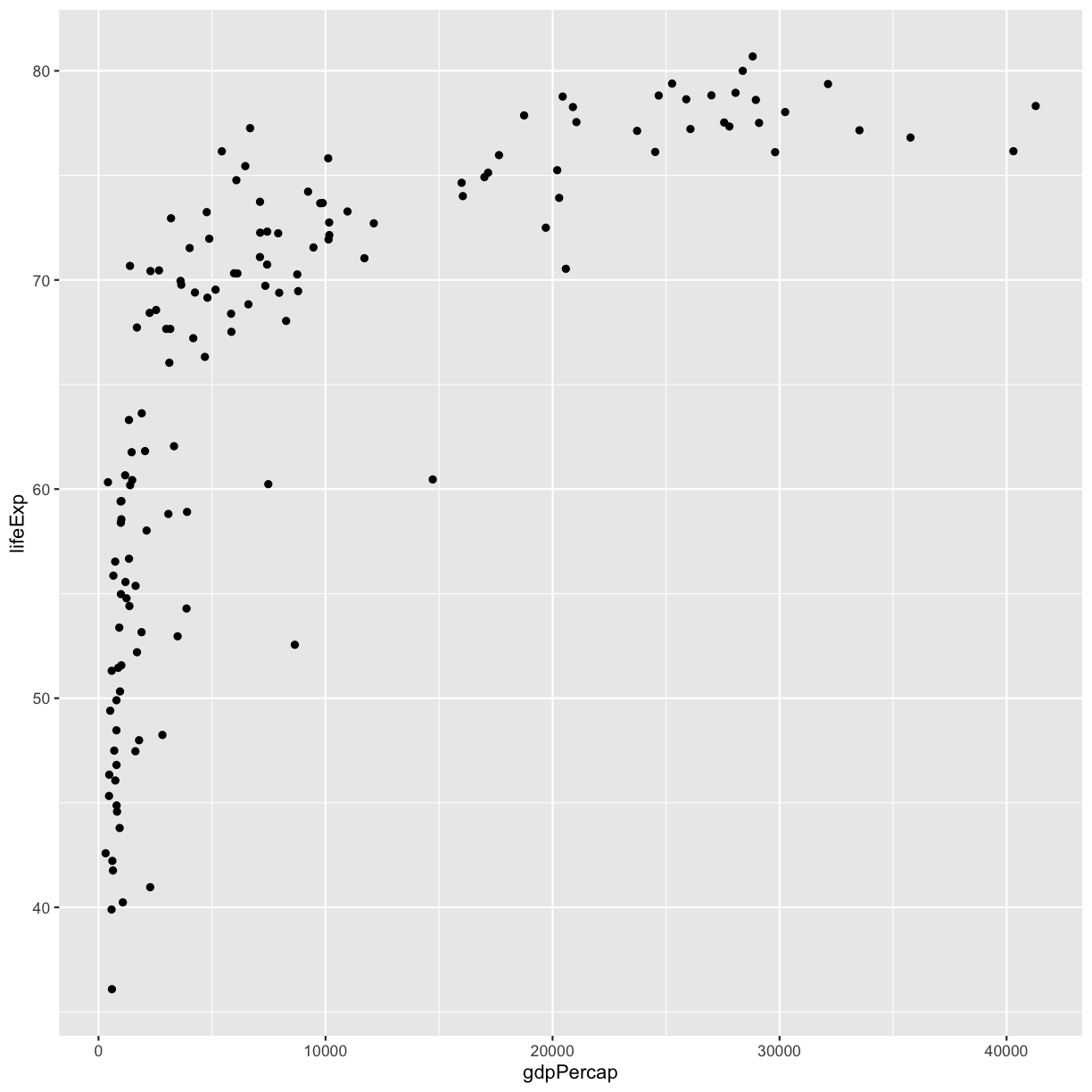
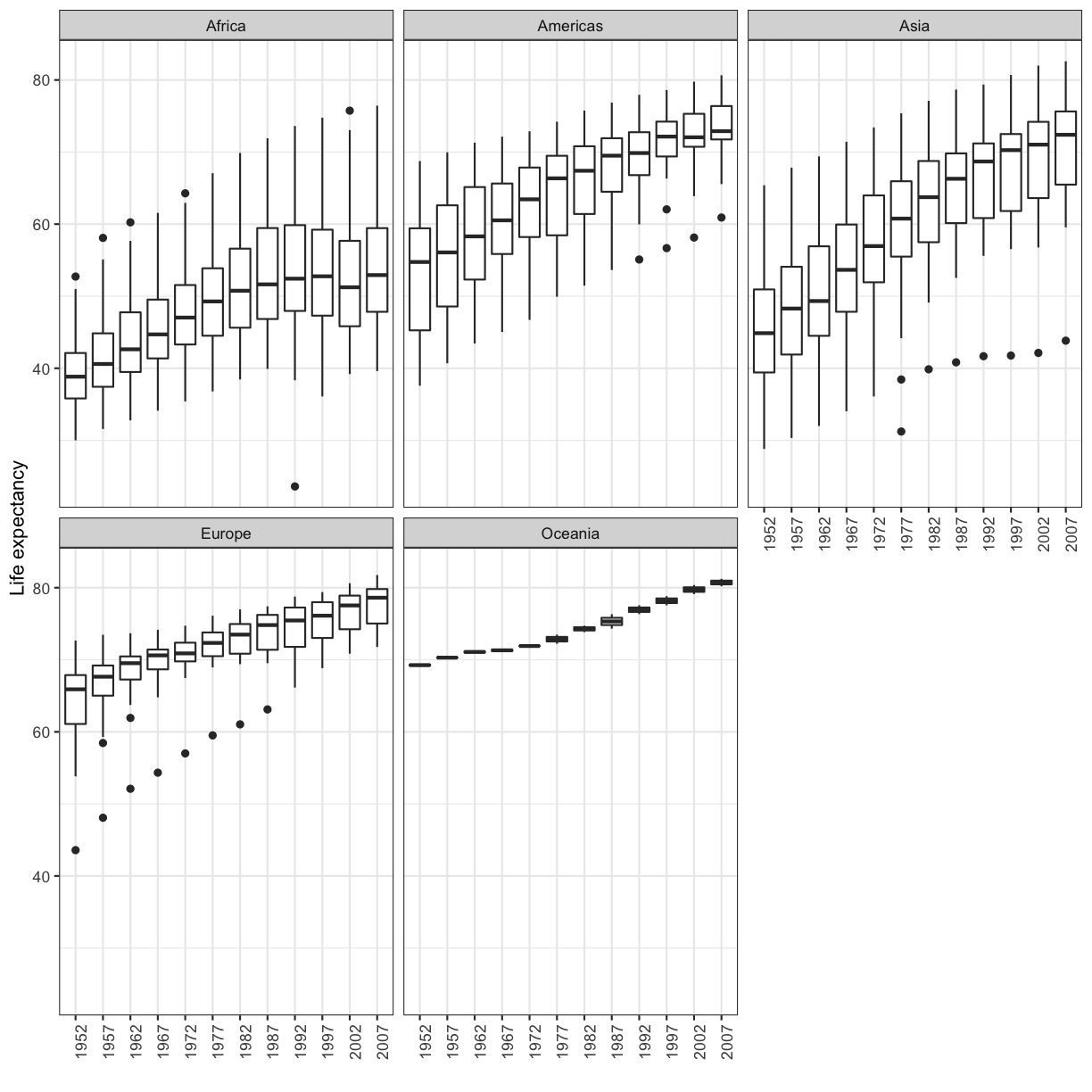
The first time we made this plot, we colored the points differently for each of the continents. This time let’s actually draw a separate box for each continent. We can do this with facet_wrap()
ggplot(gapminder_1997) +
aes(x = gdpPercap, y = lifeExp) +
geom_point() +
facet_wrap(vars(continent))
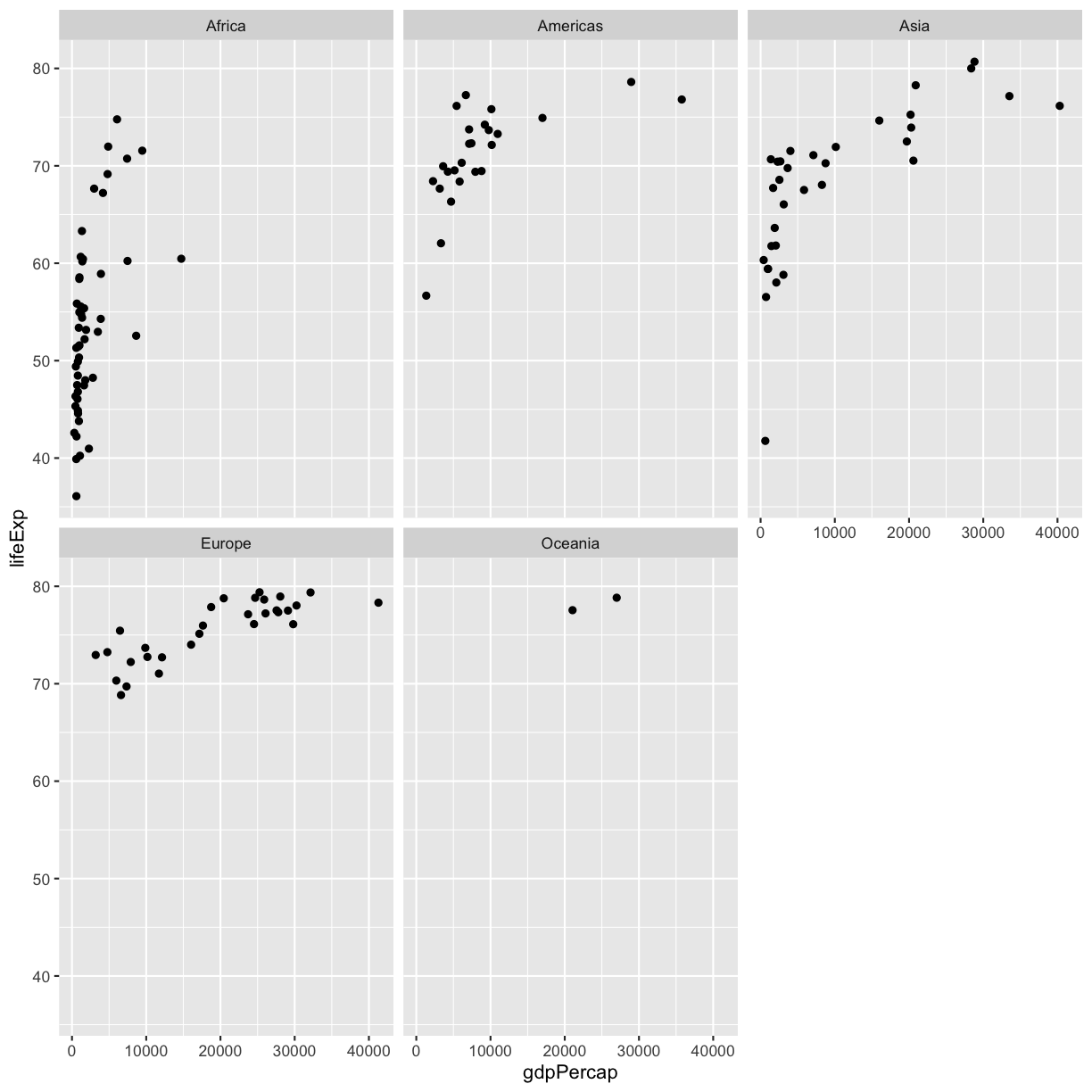 Note that
Note that facet_wrap requires this extra helper function called vars() in order to pass in the column names. It’s a lot like the aes() function, but it doesn’t require an aesthetic name. We can see in this output that we get a separate box with a label for each continent so that only the points for that continent are in that box.
The other faceting function ggplot provides is facet_grid(). The main difference is that facet_grid() will make sure all of your smaller boxes share a common axis. In this example, we will stack all the boxes on top of each other into rows so that their x axes all line up.
ggplot(gapminder_1997) +
aes(x = gdpPercap, y = lifeExp) +
geom_point() +
facet_grid(rows = vars(continent))
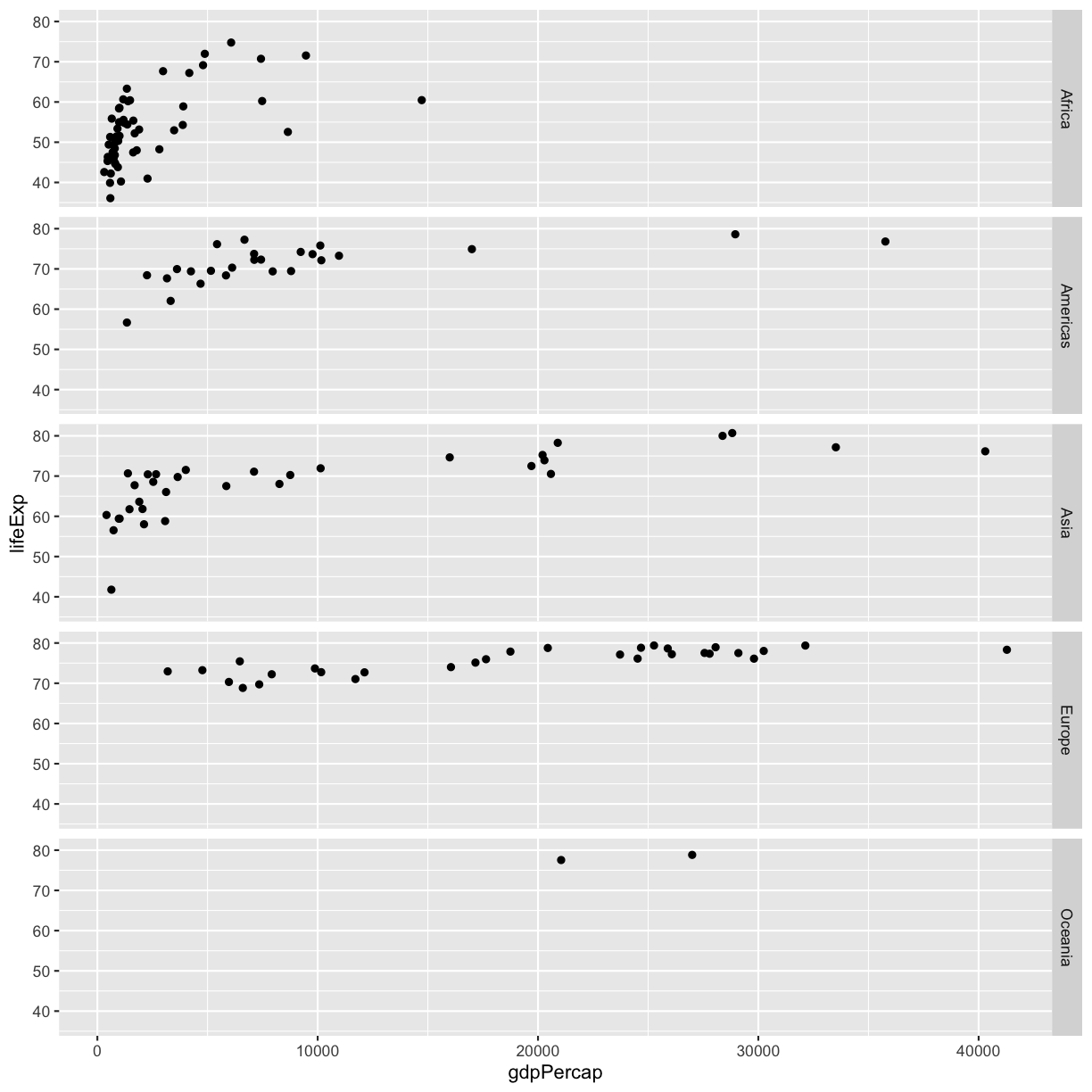
Unlike the facet_wrap output where each box got its own x and y axis, with facet_grid(), there is only one x axis along the bottom.
gh-pages
Saving plots
We’ve made a bunch of plots today, but we never talked about how to share them with your friends who aren’t running R! It’s wise to keep all the code you used to draw the plot, but sometimes you need to make a PNG or PDF version of the plot so you can share it with your PI or post it to your Instagram story.
One that’s easy if you are working in RStudio interactively is to use “Export” menu on the Plots tab. Clicking that button gives you three options “Save as Image”, “Save as PDF”, and “Copy To Clipboard”. These options will bring up a window that will let you resize and name the plot however you like.
A better option if you will be running your code as a script from the command line or just need your code to be more reproducible is to use the ggsave() function. When you call this function, it will write the last plot printed to a file in your local directory. It will determine the file type based on the name you provide. So if you call ggsave("plot.png") you’ll get a PNG file or if you call ggsave("plot.pdf") you’ll get a PDF file. By default the size will match the size of the Plots tab. To change that you can also supply width= and height= arguments. By default these values are interpreted as inches. So if you want a wide 4x6 image you could do something like:
ggsave("awesome_plot.jpg", width=6, height=4)
Exercise: Saving a plot
Try rerunning one of your plots and then saving it using
ggsave(). Find and open the plot to see if it worked!Example solution
ggplot(gapminder_1997) + aes(x = lifeExp) + geom_histogram(bins = 20)+ theme_classic()
ggsave("awesome_histogram.jpg", width=6, height=4)Check your current working directory to find the plot!
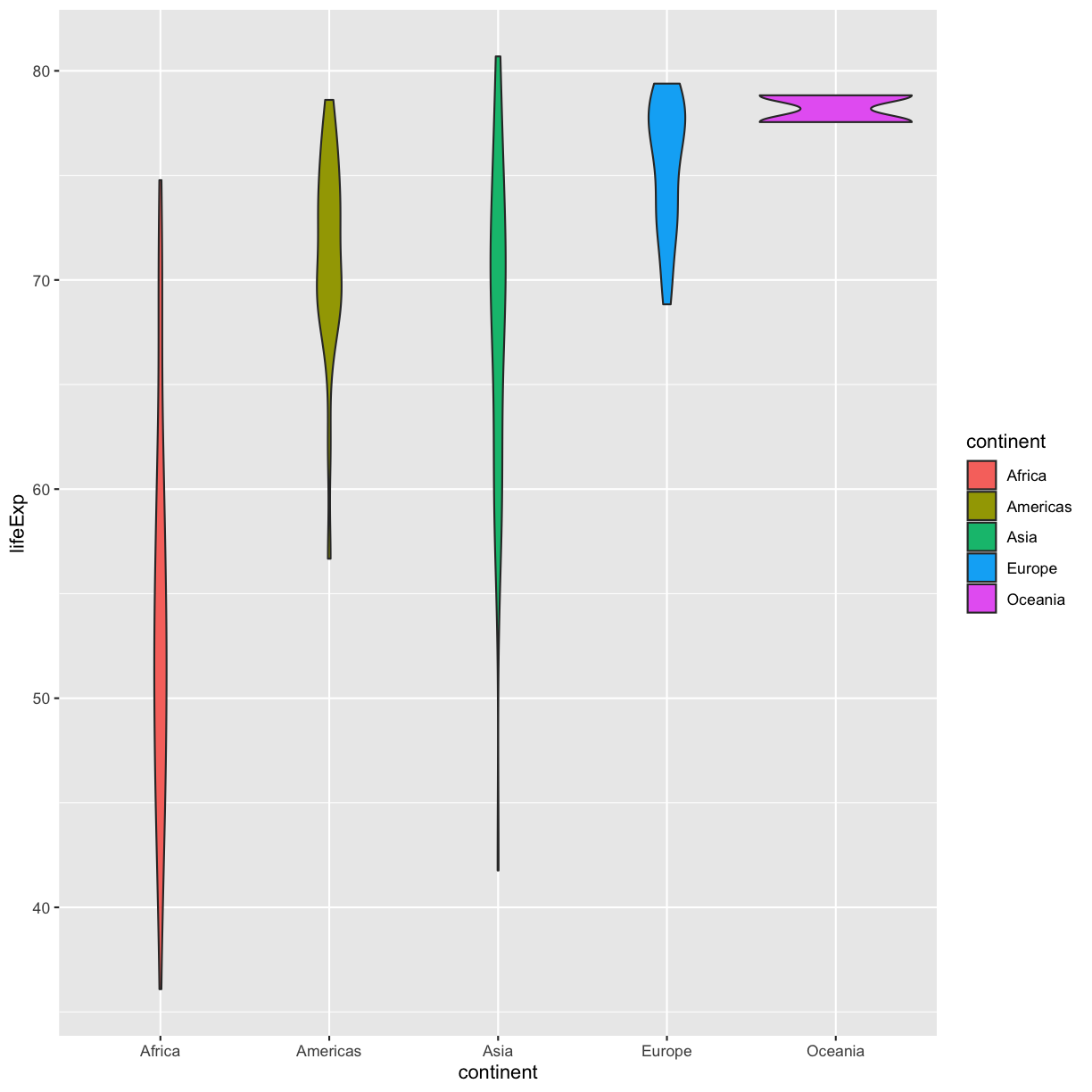
You also might want to just temporarily save a plot while you’re using R, so that you can come back to it later. Luckily, a plot is just an object, like any other object we’ve been working with! Let’s try storing our violin plot from earlier in an object called violin_plot:
violin_plot <- ggplot(data = gapminder_1997) +
aes(x = continent, y = lifeExp) +
geom_violin(aes(fill=continent))
Now if we want to see our plot again, we can just run:
violin_plot
We can also add changes to the plot. Let’s say we want our violin plot to have the black-and-white theme:
violin_plot + theme_bw()
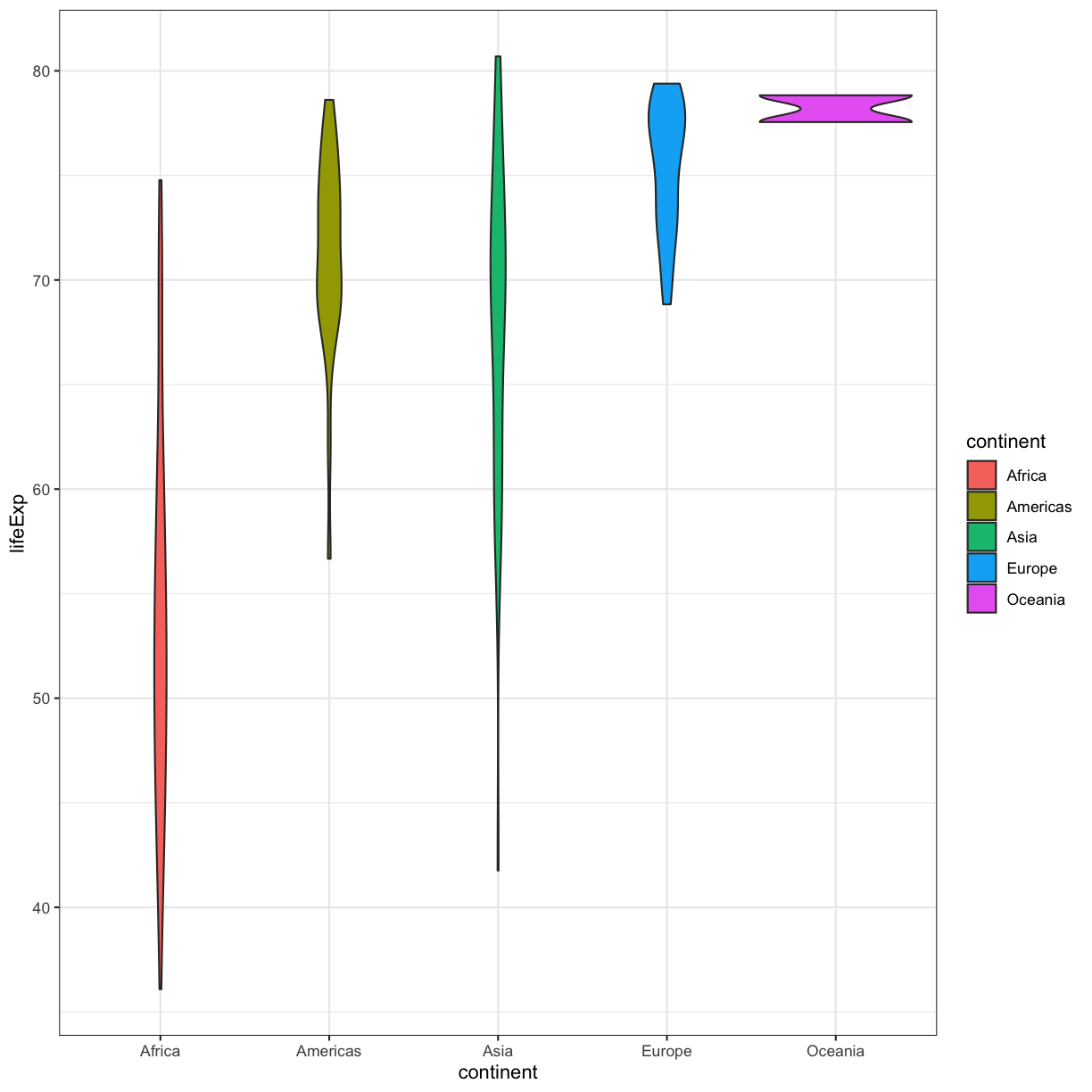
Watch out! Adding the theme does not change the violin_plot object! If we want to change the object, we need to store our changes:
violin_plot <- violin_plot + theme_bw()
We can also save any plot object we have named, even if they were not the plot that we ran most recently. We just have to tell ggsave() which plot we want to save:
ggsave("awesome_violin_plot.jpg", plot = violin_plot, width=6, height=4)
Bonus Exercise: Create and save a plot
Now try it yourself! Create your own plot using
ggplot(), store it in an object namedmy_plot, and save the plot usingggsave().Example solution
my_plot <- ggplot(data = gapminder_1997)+ aes(x = continent, y = gdpPercap)+ geom_boxplot(fill = "orange")+ theme_bw()+ labs(x = "Continent", y = "GDP Per Capita") ggsave("my_awesome_plot.jpg", plot = my_plot, width=6, height=4)
<p style="color:blue">Bonus topics!</p>
Creating complex plots
Animated plots
Sometimes it can be cool (and useful) to create animated graphs, like this famous one by Hans Rosling using the Gapminder dataset that plots GDP vs. Life Expectancy over time. Let’s try to recreate this plot!
First, we need to install and load the gganimate package, which allows us to use ggplot to create animated visuals:
install.packages(c("gganimate", "gifski"))
library(gganimate)
library(gifski)
Exercise: Reviewing how to create a scatter plot
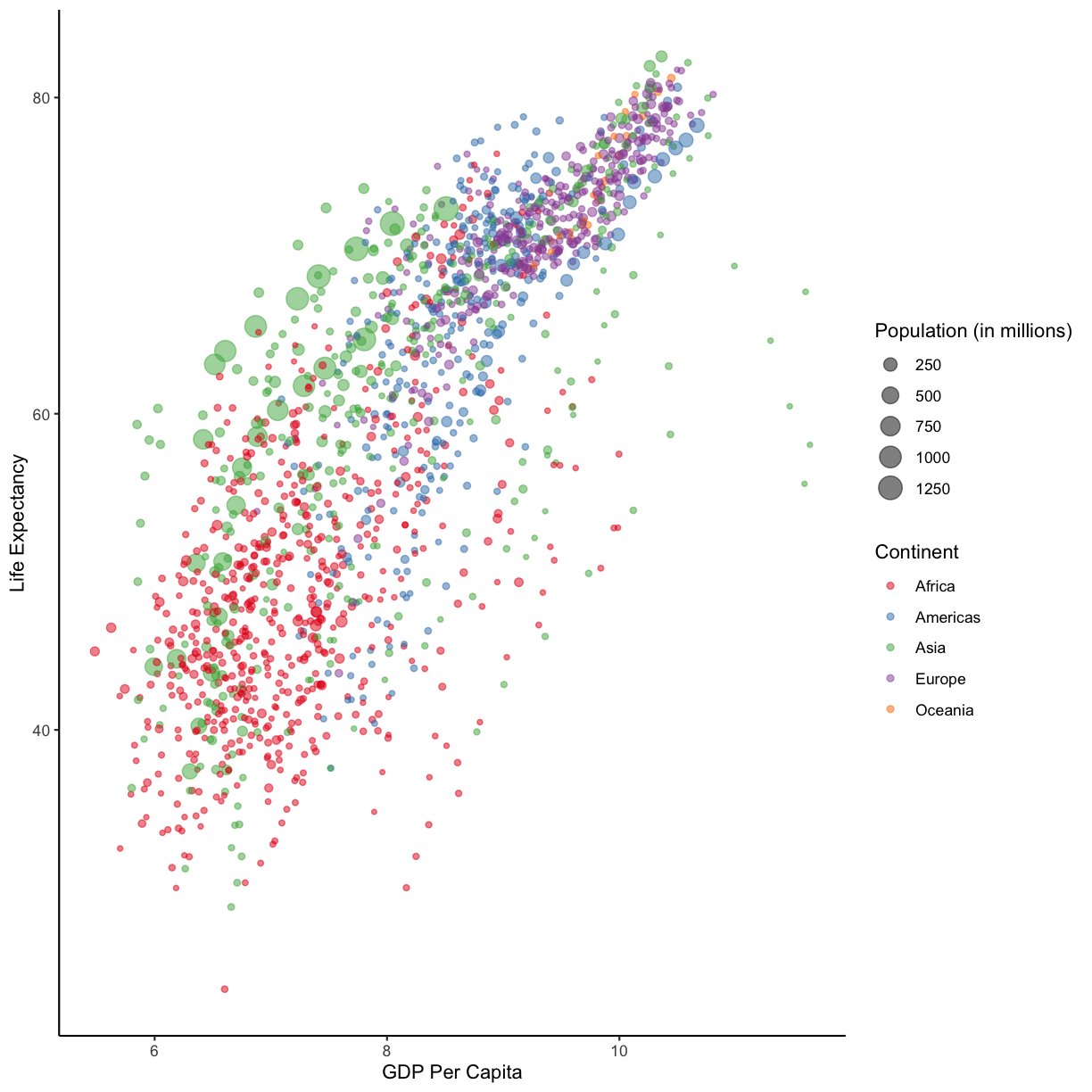
Part 1: Let’s start by creating a static plot using
ggplot(), as we’ve been doing so far. This time, lets putlog(gdpPercap)on the x-axis, to help spread out our data points, and life expectancy on our y-axis. Also map the point size to the population of the country, and the color of the points to the continent.Part 1: Let’s start by creating a static plot using
ggplot(), as we’ve been doing so far. This time let’s putlog(gdpPercap)on the x-axis, to help spread out our data points, and life expectancy on our y-axis. Also map the point size to the population of the country, and the color of the points to the continent.Solution
ggplot(data = gapminder_data)+ aes(x = log(gdpPercap), y = lifeExp, size = pop, color = continent)+ geom_point()
Part 2: Before we start to animate our plot, let’s make sure it looks pretty. Add some better axis and legend labels, change the plot theme, and otherwise fix up the plot so it looks nice. Then save the plot into an object called
staticHansPlot. When you’re ready, check out how we’ve edited our plot, below.A pretty plot (yours may look different)
staticHansPlot <- ggplot(data = gapminder_data)+ aes(x = log(gdpPercap), y = lifeExp, size = pop/1000000, color = continent)+ geom_point(alpha = 0.5) + # we made our points slightly transparent, because it makes it easier to see overlapping points scale_color_brewer(palette = "Set1") + labs(x = "GDP Per Capita", y = "Life Expectancy", color= "Continent", size="Population (in millions)")+ theme_classic() staticHansPlot
staticHansPlot <- ggplot(data = gapminder_data)+
aes(x = log(gdpPercap), y = lifeExp, size = pop/1000000, color = continent)+
geom_point(alpha = 0.5) + # we made our points slightly transparent, because it makes it easier to see overlapping points
scale_color_brewer(palette = "Set1") +
labs(x = "GDP Per Capita", y = "Life Expectancy", color= "Continent", size="Population (in millions)")+
theme_classic()
staticHansPlot
Ok, now we’re getting somewhere! But right now we’re plotting all of the years of our dataset on one plot - now we want to animate the plot so each year shows up on its own. This is where gganimate comes in! We want to add the transition_states() function to our plot.
animatedHansPlot <- staticHansPlot +
transition_states(year, transition_length = 1, state_length = 1)+
ggtitle("{closest_state}")
animatedHansPlot
Rendering [----------------------------------------------] at 3 fps ~ eta: 33s
Rendering [>-------------------------------------------] at 2.9 fps ~ eta: 34s
Rendering [>-------------------------------------------] at 2.8 fps ~ eta: 34s
Rendering [=>------------------------------------------] at 2.8 fps ~ eta: 34s
Rendering [=>------------------------------------------] at 2.9 fps ~ eta: 33s
Rendering [==>-----------------------------------------] at 2.8 fps ~ eta: 33s
Rendering [===>----------------------------------------] at 2.8 fps ~ eta: 33s
Rendering [===>----------------------------------------] at 2.8 fps ~ eta: 32s
Rendering [====>---------------------------------------] at 2.8 fps ~ eta: 32s
Rendering [====>---------------------------------------] at 2.8 fps ~ eta: 31s
Rendering [=====>--------------------------------------] at 2.8 fps ~ eta: 31s
Rendering [======>-------------------------------------] at 2.8 fps ~ eta: 30s
Rendering [=======>------------------------------------] at 2.8 fps ~ eta: 30s
Rendering [=======>------------------------------------] at 2.8 fps ~ eta: 29s
Rendering [========>-----------------------------------] at 2.8 fps ~ eta: 29s
Rendering [=========>----------------------------------] at 2.8 fps ~ eta: 28s
Rendering [==========>---------------------------------] at 2.8 fps ~ eta: 27s
Rendering [===========>--------------------------------] at 2.8 fps ~ eta: 26s
Rendering [============>-------------------------------] at 2.8 fps ~ eta: 26s
Rendering [============>-------------------------------] at 2.8 fps ~ eta: 25s
Rendering [=============>------------------------------] at 2.8 fps ~ eta: 25s
Rendering [==============>-----------------------------] at 2.8 fps ~ eta: 24s
Rendering [===============>----------------------------] at 2.8 fps ~ eta: 23s
Rendering [================>---------------------------] at 2.8 fps ~ eta: 22s
Rendering [=================>--------------------------] at 2.8 fps ~ eta: 22s
Rendering [=================>--------------------------] at 2.8 fps ~ eta: 21s
Rendering [=================>--------------------------] at 2.7 fps ~ eta: 21s
Rendering [==================>-------------------------] at 2.7 fps ~ eta: 21s
Rendering [==================>-------------------------] at 2.7 fps ~ eta: 20s
Rendering [===================>------------------------] at 2.7 fps ~ eta: 20s
Rendering [====================>-----------------------] at 2.7 fps ~ eta: 19s
Rendering [=====================>----------------------] at 2.7 fps ~ eta: 19s
Rendering [=====================>----------------------] at 2.7 fps ~ eta: 18s
Rendering [======================>---------------------] at 2.7 fps ~ eta: 18s
Rendering [======================>---------------------] at 2.7 fps ~ eta: 17s
Rendering [=======================>--------------------] at 2.7 fps ~ eta: 17s
Rendering [=======================>--------------------] at 2.7 fps ~ eta: 16s
Rendering [========================>-------------------] at 2.7 fps ~ eta: 16s
Rendering [=========================>------------------] at 2.7 fps ~ eta: 15s
Rendering [==========================>-----------------] at 2.7 fps ~ eta: 14s
Rendering [===========================>----------------] at 2.7 fps ~ eta: 14s
Rendering [===========================>----------------] at 2.7 fps ~ eta: 13s
Rendering [============================>---------------] at 2.7 fps ~ eta: 13s
Rendering [============================>---------------] at 2.7 fps ~ eta: 12s
Rendering [=============================>--------------] at 2.7 fps ~ eta: 12s
Rendering [=============================>--------------] at 2.7 fps ~ eta: 11s
Rendering [==============================>-------------] at 2.7 fps ~ eta: 11s
Rendering [===============================>------------] at 2.7 fps ~ eta: 10s
Rendering [================================>-----------] at 2.7 fps ~ eta: 10s
Rendering [================================>-----------] at 2.7 fps ~ eta: 9s
Rendering [=================================>----------] at 2.7 fps ~ eta: 9s
Rendering [=================================>----------] at 2.7 fps ~ eta: 8s
Rendering [==================================>---------] at 2.7 fps ~ eta: 8s
Rendering [==================================>---------] at 2.7 fps ~ eta: 7s
Rendering [===================================>--------] at 2.7 fps ~ eta: 7s
Rendering [====================================>-------] at 2.7 fps ~ eta: 6s
Rendering [=====================================>------] at 2.7 fps ~ eta: 5s
Rendering [======================================>-----] at 2.7 fps ~ eta: 4s
Rendering [=======================================>----] at 2.7 fps ~ eta: 4s
Rendering [=======================================>----] at 2.7 fps ~ eta: 3s
Rendering [========================================>---] at 2.7 fps ~ eta: 3s
Rendering [========================================>---] at 2.7 fps ~ eta: 2s
Rendering [=========================================>--] at 2.7 fps ~ eta: 2s
Rendering [=========================================>--] at 2.7 fps ~ eta: 1s
Rendering [==========================================>-] at 2.7 fps ~ eta: 1s
Rendering [===========================================>] at 2.7 fps ~ eta: 0s
Rendering [============================================] at 2.7 fps ~ eta: 0s

Awesome! This is looking sweet! Let’s make sure we understand the code above:
- The first argument of the
transition_states()function tellsggplot()which variable should be different in each frame of our animation: in this case, we want each frame to be a differentyear. - The
transition_lengthandstate_lengtharguments are just some of thegganimatearguments you can use to adjust how the animation progresses from one frame to the next. Feel free to play around with those parameters, to see how they affect the animation (or check out moregganmiateoptions here!). - Finally, we want the title of our plot to tell us which year our animation is currently showing. Using “{closest_state}” as our title allows the title of our plot to show which
yearis currently being plotted.
So we’ve made this cool animated plot - how do we save it? For gganimate objects, we can use the anim_save() function. It works just like ggsave(), but for animated objects.
anim_save("hansAnimatedPlot.gif",
plot = animatedHansPlot,
renderer = gifski_renderer())
Map plots
The ggplot library also has useful functions to draw your data on a map. There are lots of different ways to draw maps but here’s a quick example using the gampminder data. Here we will plot each country with a color indicating the life expectancy in 1997.
# make sure names of countries match between the map info and the data
mapdata <- map_data("world") %>%
mutate(region = recode(region,
USA="United States",
UK="United Kingdom"))
#install.packages("mapproj")
gapminder_1997 %>%
ggplot() +
geom_map(aes(map_id=country, fill=lifeExp), map=mapdata) +
expand_limits(x = mapdata$long, y = mapdata$lat) +
coord_map(projection = "mollweide", xlim = c(-180, 180)) +
ggthemes::theme_map()
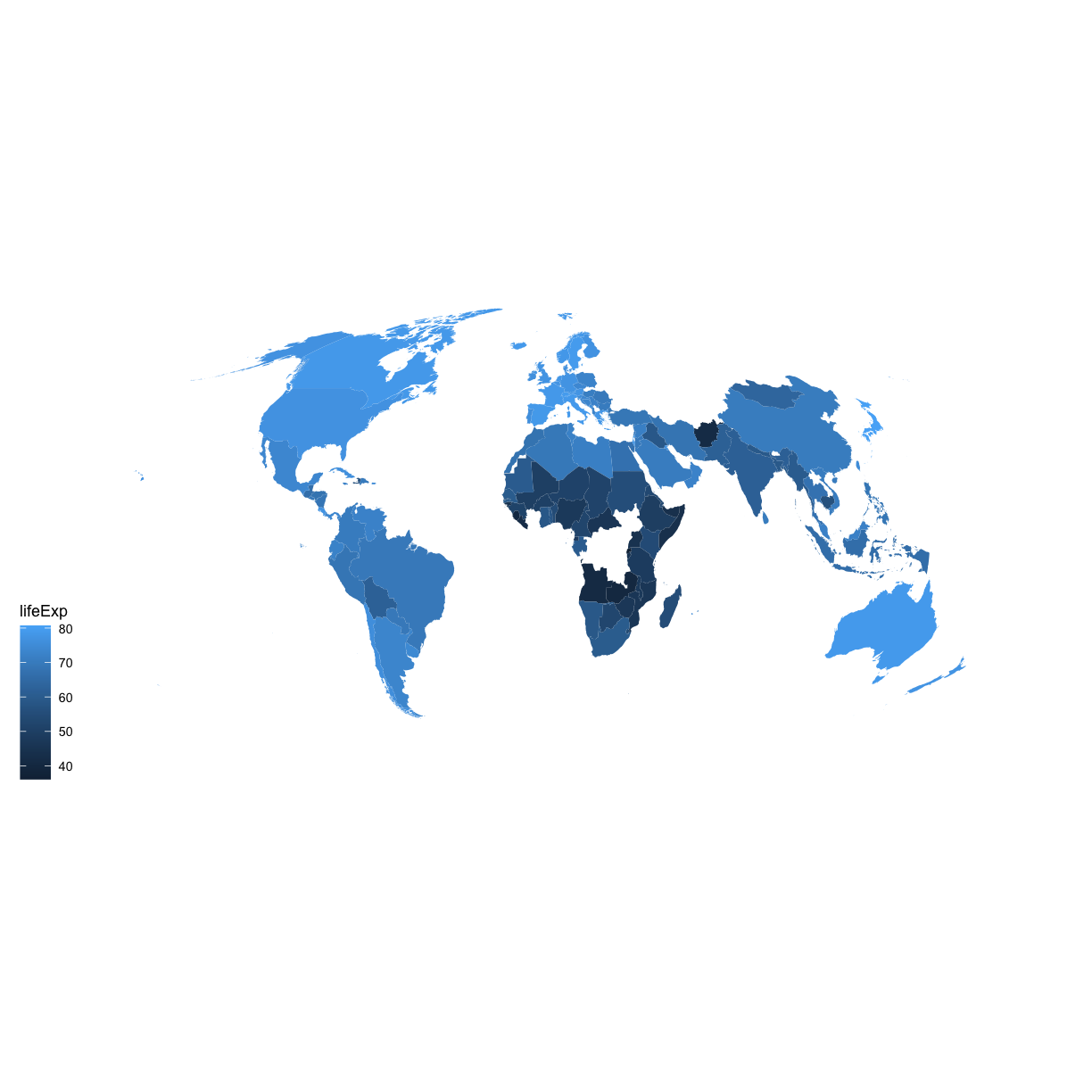
Notice that this map helps to show that we actually have some gaps in the data. We are missing observations for counties like Russia and many countries in central Africa. Thus, it’s important to acknowledge that any patterns or trends we see in the data might not apply to those regions.
Glossary of terms
- Aesthetic: a visual property of the objects (geoms) drawn in your plot (like x position, y position, color, size, etc)
- Aesthetic mapping (aes): This is how we connect a visual property of the plot to a column of our data
- Comments: lines of text in our code after a
#that are ignored (not evaluated) by R - Geometry (geom): this describes the things that are actually drawn on the plot (like points or lines)
- Facets: Dividing your data into non-overlapping groups and making a small plot for each subgroup
- Layer: Each ggplot is made up of one or more layers. Each layer contains one geometry and may also contain custom aesthetic mappings and private data
- Factor: a way of storing data to let R know the values are discrete so they get special treatment
Key Points
R is a free programming language used by many for reproducible data analysis.
Use
read_csvto read tabular data in R.Geometries are the visual elements drawn on data visualizations (lines, points, etc.), and aesthetics are the visual properties of those geometries (color, position, etc.).
Use
ggplot()and geoms to create data visualizations, and save them usingggsave().
The Unix Shell
Overview
Teaching: 60 min
Exercises: 15 minQuestions
What is a command shell and why would I use one?
How can I move around on my computer?
How can I see what files and directories I have?
How can I specify the location of a file or directory on my computer?
How can I create, copy, and delete files and directories?
How can I edit files?
Objectives
Explain how the shell relates to users’ programs.
Explain when and why command-line interfaces should be used instead of graphical interfaces.
Construct absolute and relative paths that identify specific files and directories.
Demonstrate the use of tab completion and explain its advantages.
Create a directory hierarchy that matches a given diagram.
Create files in the directory hierarchy using an editor or by copying and renaming existing files.
Delete, copy, and move specified files and/or directories.
Contents
Introducing the Shell
Motivation
Usually you move around your computer and run programs through graphical user interfaces (GUIs). For example, Finder for Mac and Explorer for Windows. These GUIs are convenient because you can use your mouse to navigate to different folders and open different files. However, there are some things you simply can’t do from these GUIs.
The Unix Shell (or the command line) allows you to do everything you would do through Finder/Explorer, and a lot more. But it’s so scary! I thought so at first, too. Since then, I’ve learned that it’s just another way to navigate your computer and run programs, and it can be super useful for your work. For instance, you can use it to combine existing tools into a pipeline to automate analyses, you can write a script to do things for you and improve reproducibility, you can interact with remote machines and supercomputers that are far away from you, and sometimes it’s the only option for the program you want to run.
We’re going to use it to:
- Organize our R code and plots from the R plotting lesson.
- Perform version control using git during the rest of the workshop.
What the Shell looks like
When you open up the terminal for the first time, it can look pretty scary - it’s basically just a blank screen. Don’t worry - we’ll take you through how to use it step by step.
The first line of the shell shows a prompt - the shell is waiting for an input. When you’re following along in the lesson, don’t type the prompt when typing commands. To make the prompt the same for all of us, run these commands:
PS1_A=$PS1
PS1='$ '
#PS1=$PS1_A # Remove the leading "#" and execute this line to restore your original prompt.
Tree Structure
The first thing we need to learn when using the shell is how to get around our computer.
The shell folder (directory) structure is the same file structure as you’re used to.
We call the way that different directories are nested the “directory tree”.
You start at the root directory (/) and you can move “up” and “down” the tree. Here’s an example:
Now that we understand directory trees a bit, let’s check it out from the command line.
We can see where we are by using the command pwd which stands for “print working directory”, or the directory we are currently in:
pwd
/home/USERNAME/
Congrats! You just ran your first command from the command line. The output is a file path to a location (a directory) on your computer.
The output will look a little different depending on what operating system you’re using:
- Mac:
/Users/USERNAME - Linux:
/home/USERNAME - Windows:
/c/Users/USERNAME
Let’s check to see what’s in your home directory using the ls command, which lists all of the files in your working directory:
ls
Desktop Downloads Movies Pictures
Documents Library Music Public
You should see some files and directories you’re familiar with such as Documents and Desktop.
If you make a typo, don’t worry. If the shell can’t find a command you type, it will show you a helpful error message.
ks
ks: command not found
This error message tells us the command we tried to run, ks, is not a command that is recognized, letting us know we might have made a mistake when typing.
If you ever want to cancel a command and get a fresh prompt, you can hold your control key and hit c.
control-c is also how you can stop a running program.
Also, you can use the command clear to clear your screen and get a fresh prompt.
Man and Help
Now that we know how to list files with ls, we can learn how to look up the manual pages for unix shell commands. If you want to learn more about a command we can use man to look up its manual page. which will open with ls. We can navigate the man page to view the description of a command and its options. For example, if you want to know more about the navigation options of ls you can type man ls on the command line.
man ls
On the manual page for ls, we see a section titled options. These options, also called flags, allow us to customize how ls runs.
One very helpful flag that is available for any command is -h or --help which will print brief documentation for the command.
man -h
man --help
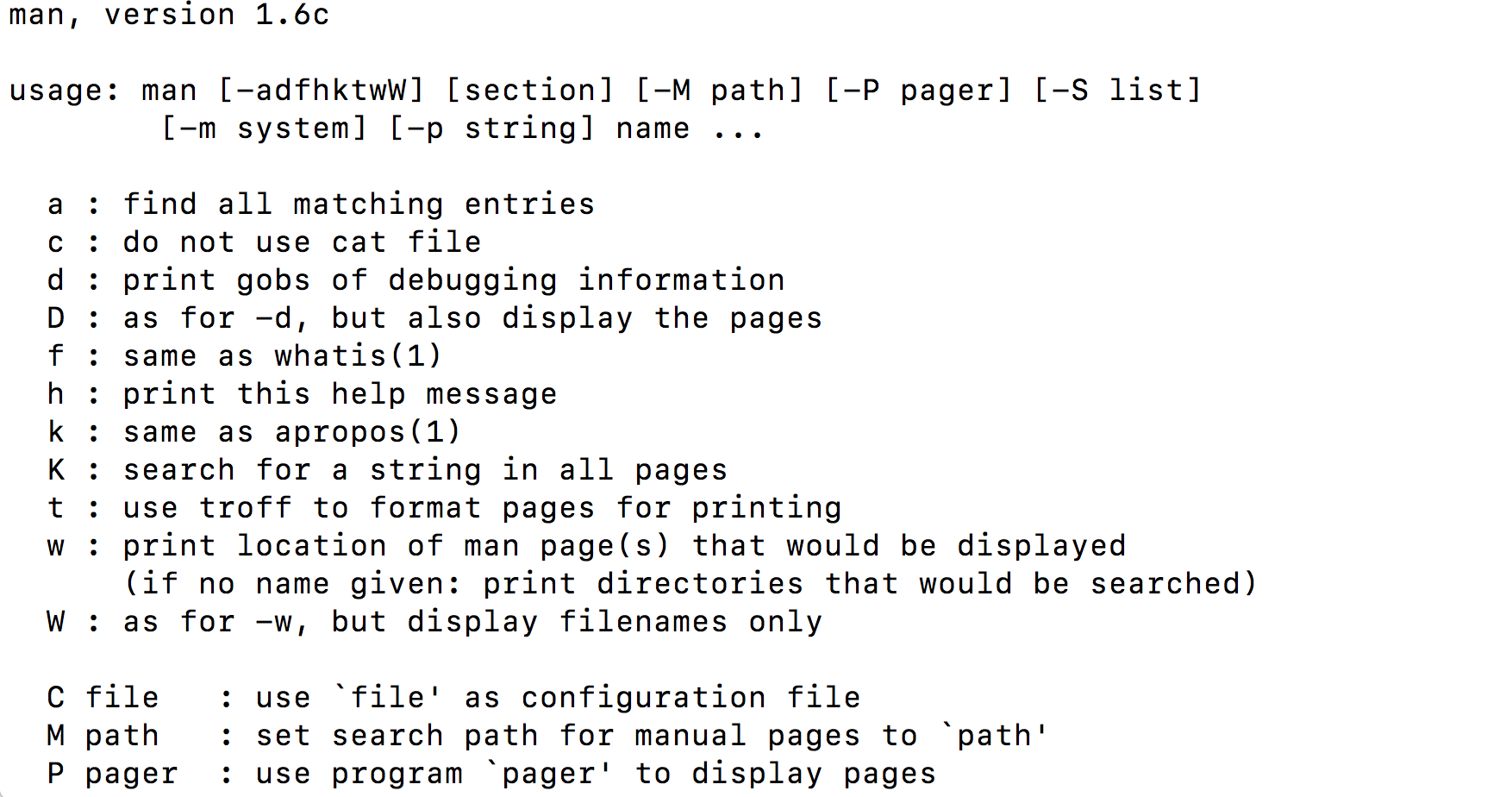
Using the Manual Pages
Use
manto open the manual for the commandls.What flags would you use to…
- Print files in order of size?
- Print files in order of the last time they were edited?
- Print hidden files (files that begin with
.)?- Print more information about the files?
- Print more information about the files with unit suffixes?
- Print files in order of size AND also print more information about the files?
Solution
ls -Sls -tls -als -lls -lhls -lS
Next, let’s move to our Desktop. To do this, we use cd to change directories.
Run the following command:
cd Desktop
Let’s see if we’re in the right place:
pwd
/home/USERNAME/Desktop
We just moved down the directory tree into the Desktop directory.
What files and directories do you have on your Desktop? How can you check?
ls
list.txt
un-report
notes.pdf
Untitled.png
Your Desktop will likely look different, but the important thing is that you see the folder we worked in for the R plotting lesson.
Is the un-report directory listed on your Desktop?
How can we get into the un-report directory?
cd un-report
We just went down the directory tree again.
Let’s see what files are in un-report; add the -1 (that’s the number one) to
format the listing as a single column of files:
ls -1
awesome_plot.jpg
awesome_violin_plot.jpg
gapminder_1997.csv
gapminder_data.csv
gdp_population.R
Is it what you expect? Are the files you made in the R plotting lesson there?
Now let’s move back up the directory tree. First, let’s try this command:
cd Desktop
cd: Desktop: No such file or directory
This doesn’t work because the Desktop directory is not within the directory that we are currently in.
To move up the directory tree, you can use .., which is the parent of the current directory:
cd ..
pwd
/home/USERNAME/Desktop
Everything that we’ve been doing is working with file paths. We tell the computer where we want to go using cd plus the file path. We can also tell the computer what files we want to list by giving a file path to ls:
ls un-report
awesome_plot.jpg
awesome_violin_plot.jpg
gapminder_1997.csv
gapminder_data.csv
gdp_population.R
ls ..
list.txt
un-report
notes.pdf
Untitled.png
What happens if you just type cd without a file path?
cd
pwd
/home/USERNAME
It takes you back to your home directory!
To get back to your projects directory you can use the following command:
cd Desktop/un-report
An extremely powerful technique that will save tons of time is using tab completion. As an example, go back to your home dir:
cdNow type
cdfollowed by a space andDeand immediately hit thetabkey. The Bash shell will find files/directories in the working directory that start withDe. If it finds a match, it will enter the match on your command line.cd Desktop/But don’t hit
Enter/Returnyet - instead typeunimmediately hit thetabkey. The Bash shell should autocomplete to thiscd Desktop/un-reportNow hit enter to execute the command to change to the new directory.
Another very powerful technique is Bash history. Use the up-arrow and down-arrow keys to review the last several commands. If you find a command to re-execute, you can simply press
Enter/Return.Bash history and tab-completion can save a lot of time and frustration - use them liberally!
We have been using relative paths, meaning you use your current working directory to get to where you want to go.
You can also use the absolute path, or the entire path from the root directory. What’s listed when you use the pwd command is the absolute path:
pwd
You can also use ~ for the path to your home directory:
cd ~
pwd
/home/USERNAME
Absolute vs Relative Paths
Starting from
/Users/amanda/data, which of the following commands could Amanda use to navigate to her home directory, which is/Users/amanda?
cd .cd /cd /home/amandacd ../..cd ~cd homecd ~/data/..cdcd ..Solution
- No:
.stands for the current directory.- No:
/stands for the root directory.- No: Amanda’s home directory is
/Users/amanda.- No: this goes up two levels, i.e. ends in
/Users.- Yes:
~stands for the user’s home directory, in this case/Users/amanda.- No: this would navigate into a directory
homein the current directory if it exists.- Yes: unnecessarily complicated, but correct.
- Yes: shortcut to go back to the user’s home directory.
- Yes: goes up one level.
Working with files and directories
Now that we know how to move around your computer using the command line, our next step is to organize the project that we started in the R plotting lesson You might ask: why would I use the command line when I could just use the GUI? My best response is that if you ever need to use a high-performance computing cluster (such as Great Lakes at the University of Michigan), you’ll have no other option. You might also come to like it more than clicking around to get places once you get comfortable, because it’s a lot faster!
First, let’s make sure we’re in the right directory (the un-reports directory):
pwd
/home/USERNAME/Desktop/un-reports
If you’re not there, cd to the correct place.
Next, let’s remind ourselves what files are in this directory:
ls
awesome_plot.jpg
awesome_violin_plot.jpg
gapminder_1997.csv
gapminder_data.csv
gdp_population.R
You can see that right now all of our files are in our main directory. However, it can start to get crazy if you have too many different files of different types all in one place! We’re going to create a better project directory structure that will help us organize our files. This is really important, particularly for larger projects. If you’re interested in learning more about structuring computational biology projects in particular, here is a useful article.
What do you think good would be a good way to organize our files?
One way is the following:
.
├── code
│ └── gdp_population.R
├── data
│ ├── gapminder_1997.csv
└── gapminder_data.csv
└── figures
├── awesome_plot.jpg
└── awesome_violin_plot.jpg
The R script goes in the code directory, the gapminder datasets go in the data directory, and the figures go in the figures directory. This way, all of the files are organized into a clearer overall structure.
A few notes about naming files and directories:
- Don’t use whitespaces because they’re used to break arguments on the command line, so it makes things like moving and viewing files more complicated.
Instead you can use a dash (
-) or an underscore (_). - Don’t start names with a dash (
-) because the shell will interpret it incorrectly. - Stick with letters, numbers, periods, dashes, and underscores, because other symbols (e.g.
^,&) have special meanings. - If you have to refer to names of files or directories with whitespace or other special characters, use double quotes. For example, if you wanted to change into a directory called
My Code, you will want to typecd "My Code", notcd My Code. - Unfortunately, how upper and lower case is treated can differ across systems. Advice: assume that filesystems are case sensitive but never use case alone to differentiate two files.
So how do we make our directory structure look like this?
First, we need to make a new directory. Let’s start with the code directory. To do this, we use the command mkdir plus the name of the directory we want to make:
mkdir code
Now, let’s see if that directory exists now:
ls
awesome_plot.jpg
awesome_violin_plot.jpg
code
gapminder_1997.csv
gapminder_data.csv
gdp_population.R
How can we check to see if there’s anything in the code directory?
ls code
Nothing in there yet, which is expected since we just made the directory.
The next step is to move the .R file into the code directory. To do this, we use the mv command. The first argument after mv is the file you want to move, and the second argument is the place you want to move it:
mv gdp_population.R code
Okay, let’s see what’s in our current directory now:
ls
awesome_plot.jpg
awesome_violin_plot.jpg
code
gapminder_1997.csv
gapminder_data.csv
gdp_population.R is no longer there! Where did it go? Let’s check the code directory, where we moved it to:
ls code
gdp_population.R
There it is!
Okay, now we have the code and data in the right place. But we have several figures that should still be in their own directory.
First, let’s make a figures directory:
mkdir figures
Next, we have to move the figures. But we have so many figures! It’d be annoying to move them one at a time. Thankfully, we can use a wildcard to move them all at once. Wildcards are used to match files and directories to patterns.
One example of a wildcard is the asterisk, *. This special character is interpreted as “multiple characters of any kind”.
Let’s see how we can use a wildcard to list only files with the extension .jpg:
ls *jpg
awesome_plot.jpg
awesome_violin_plot.jpg
See how only the files ending in .jpg were listed? The shell expands the wildcard to create a list of matching file names before running the commands. Can you guess how we move all of these files at once to the figures directory?
mv *jpg figures
Working with Wildcards
Suppose we are in a directory containing the following files:
cubane.pdb ethane.pdb methane.pdb octane.pdb pentane.pdb propane.pdb README.mdWhat would be the output of the following commands?
ls *ls *.pdbls *ethane.pdbls *anels p*Solution
cubane.pdb ethane.pdb methane.pdb octane.pdb pentane.pdb propane.pdb README.mdcubane.pdb ethane.pdb methane.pdb octane.pdb pentane.pdb propane.pdbethane.pdb methane.pdb- None. None of the files end in only
ane. This would have listed files ifls *ane*were used instead.pentane.pdb propane.pdb
Creating directories and moving files
Create a
datadirectory and movegapminder_data.csvandgapminder_1997.csvinto the newly createddatadirectory.Solution
From the
un-reportdirectory:mkdir data mv gapminder_data.csv data mv gapminder_1997.csv data
We can also use the wildcard to list all of the files in all of the directories:
ls *
code:
gdp_population.R
data:
gapminder_1997.csv gapminder_data.csv
figures:
awesome_plot.jpg awesome_violin_plot.jpg
This output shows each directory name, followed by its contents on the next line. As you can see, all of the files are now in the right place!
Let’s look more closely at the un-report directory; first run this command:
ls -1 -F
code/
data/
figures/
Now use arrow-up and add an extra option -a to show hidden files:
ls -1 -F -a
./
../
.Rproj.user
code/
data/
figures/
Hidden files begin with a .; by default they are not displayed in ls but
otherwise they are just like other files. Note above that you can see ./ and
../ which reference the current directory and parent directory respectively.
You can also see .Rproj.user which is a hidden file used by R-Studio.
In addition to moving files, we sometimes need to create and delete them. We’ll
illustrate this process in a new directory named temp.
cd ~/Desktop/un-report
mkdir temp
cd temp
pwd
~/Desktop/un-report/temp
The cp command copies files from one location (source) to another (destination).
cp ../data/gapminder_1997.csv gapminder_1997_copy.csv
ls
gapminder_1997_copy.csv
The rm command removes (deletes) a file. Be very careful with this command
because it is permanent and there is often no way to recover a deleted file.
rm gapminder_1997_copy.csv
cd ..
ls temp
You can remove an empty directory with the rmdir command:
rmdir temp
ls temp
ls: temp: No such file or directory
Viewing Files
The command cat (short for concatenate) prints the entire contents of a file
to the terminal.
cat data/gapminder_1997.csv
The command head shows the first 10 lines of a file.
head data/gapminder_1997.csv
country,pop,continent,lifeExp,gdpPercap
Afghanistan,22227415,Asia,41.763,635.341351
Albania,3428038,Europe,72.95,3193.054604
Algeria,29072015,Africa,69.152,4797.295051
Angola,9875024,Africa,40.963,2277.140884
Argentina,36203463,Americas,73.275,10967.28195
Australia,18565243,Oceania,78.83,26997.93657
Austria,8069876,Europe,77.51,29095.92066
Bahrain,598561,Asia,73.925,20292.01679
Bangladesh,123315288,Asia,59.412,972.7700352
You can adjust the number of lines returned by head:
head -n 3 data/gapminder_1997.csv
country,pop,continent,lifeExp,gdpPercap
Afghanistan,22227415,Asia,41.763,635.341351
Albania,3428038,Europe,72.95,3193.054604
To view and navigate the contents of a file we can use the command less. This will open a full screen view of the file.
Here is what we should expect to see when running the command less on our gapminder_data.csv file:
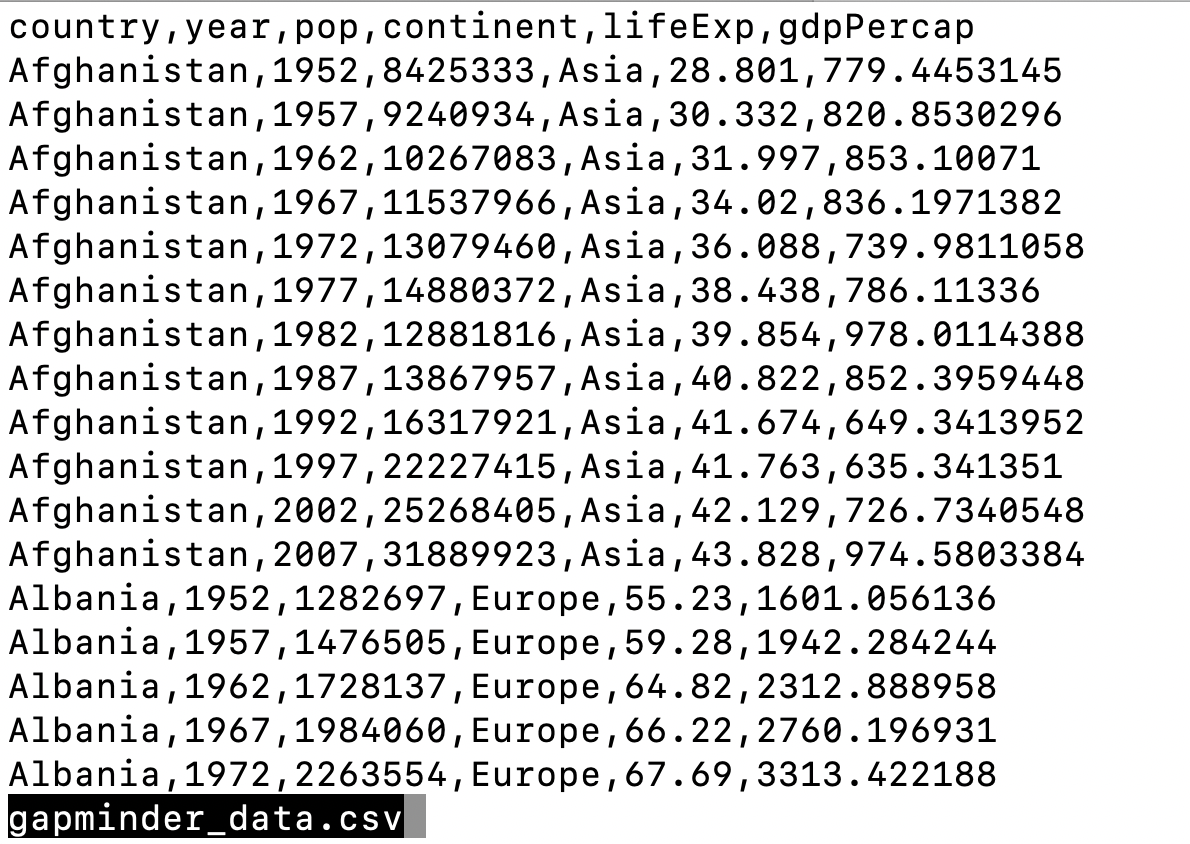
To navigate, press spacebar to scroll to the next page and b to scroll up to the previous page. You can also use the up and down arrows to scroll line-by-line. Note that less defaults to line wrapping, meaning that any lines longer than the width of the screen will be wrapped to the next line, (to disable this use the option -S when running less, ex less -S file.txt). To exit less, press the letter q.
We should note that not all file types can be viewed with less. While we can open PDFs and excel spreadsheets easily with programs on our computer, less doesn’t render them well on the command line. For example, if we try to less a .pdf file we will see a warning.
less file.pdf
file.pdf may be a binary file. See it anyway?
If we say “yes”, less will render the file but it will appear as a seemingly random display of characters that wont make much sense to us.

Sometimes, commands will have multiple flags that we want to use at the same time.
less -M -N -S [FILE]
or equivalently
less -MNS [FILE]
Decoding options
Execute
less -MNS data/gapminder_1997.csv. What are some ways you could understand what each of the three options in this command are doing?Solutions
- Compare above with
less data/gapminder_1997.csv- Run each flag individually and iteratively compare with
less data/gapminder_1997.csvman less- https://explainshell.com/
Editing Files
Beyond viewing the content of files, we may want to be able to edit or write files on the command line. There are many different text editors you can use to edit files on the command line, but we will talk about nano since it is a bit easier to learn. To edit a file with nano type nano file.txt. If the file exists, it will open the file in a nano window, if the file does not exist it will be created. One nice feature of nano is that it has a cheat sheet along the bottom with some common commands you’ll need. When you are ready to save (write) your file, you type Ctrl+O. Along the bottom will appear a prompt for the file name to write to. The current name of the file will appear here, to keep the name as it is hit enter otherwise you can change the name of the file then hit enter. To exit nano, press Ctrl+X. If you forget to save before exiting, no worries nano will prompt you to first save the file.
Since we moved around files when we organized our project directory we will have to update our R script. The path we use to read in our dataset is no longer correct. We will use nano to update the path to our new directory structure.
nano code/gdp_population.R
gapminder_data <- read_csv("data/gapminder_data.csv")
Great! Now as an exercise we can change the paths to write out figures.
Editing file paths with nano
Use nano to edit the file paths of the figures saved in
code/gdp_population.Rto match our new directory structure.Solution
nano code/gdp_population.REdit the lines in
code/gdp_population.Rwhere plots are saved:ggsave("figures/awesome_plot.jpg", width=6, height=4) ggsave("figures/awesome_histogram.jpg", width=6, height=4)
History
Earlier we mentioned that the up-arrow and down-arrow allow you to interactively
scroll through your history of commands. Another way to repeat previous work is
to use the history command to get a list of the last few hundred commands that
have been executed, and then to use !123 (where ‘123’ is replaced by the
command number) to repeat one of those commands. For example, if you type this:
history | tail
10 less figures/awesome_plot.jpg
11 less data/gapminder_data.csv
12 less -MNS data/gapminder_data.csv
13 nano code/gdp_population.R
14 history
then you can re-run
less -MNS data/gapminder_data.csvsimply by typing!12.
Glossary of terms
- root: the very top of the file system tree
- absolute path: the location of a specific file or directory starting from the root of the file system tree
-
relative path: the location of a specific file or directory starting from where you currently are in the file system tree
pwd: Print working directory - prints the absolute path from the root directory to the directory where you currently are.ls: List files - lists files in the current directory. You can provide a path to list files to another directory as well (ls [path]).cd [path]: Change directories - move to another folder.mkdir: Make directory - creates a new directory..: This will move you up one level in the file system treemv: Move - move a file to a new location (mv [file] [/path/to/new/location]) OR remaning a file (mv [oldfilename] [newfilename])less: - quick way to view a document without using a full text editorman: Manual - allows you to view the bash manual for another command (e.g.man ls)-h/--help: Help - argument that pulls up the help manual for a programnano: a user-friendly text editor*: Wildcard - matches zero of more characters in a filename
Resources
- https://www.kernel.org/doc/man-pages/
- https://explainshell.com/
- https://files.fosswire.com/2007/08/fwunixref.pdf
- http://www.mathcs.emory.edu/~valerie/courses/fall10/155/resources/unix_cheatsheet.html
Key Points
A shell is a program whose primary purpose is to read commands and run other programs.
Tab completion can help you save a lot of time and frustration.
The shell’s main advantages are its support for automating repetitive tasks and its capacity to access network machines.
Information is stored in files, which are stored in directories (folders).
Directories nested in other directories for a directory tree.
cd [path]changes the current working directory.
ls [path]prints a listing of a specific file or directory.
lslists the current working directory.
pwdprints the user’s current working directory.
/is the root directory of the whole file system.A relative path specifies a location starting from the current location.
An absolute path specifies a location from the root of the file system.
Directory names in a path are separated with
/on Unix, but\on Windows.
..means ‘the directory above the current one’;.on its own means ‘the current directory’.
cp [old] [new]copies a file.
mkdir [path]creates a new directory.
mv [old] [new]moves (renames) a file or directory.
rm [path]removes (deletes) a file.
*matches zero or more characters in a filename.The shell does not have a trash bin — once something is deleted, it’s really gone.
Intro to Git & GitHub
Overview
Teaching: 90 min
Exercises: 30 minQuestions
What is version control and why should I use it?
How do I get set up to use Git?
How do I share my changes with others on the web?
How can I use version control to collaborate with other people?
Objectives
Explain what version control is and why it’s useful.
Configure
gitthe first time it is used on a computer.Learn the basic git workflow.
Push, pull, or clone a remote repository.
Describe the basic collaborative workflow with GitHub.
Contents
- Background
- Setting up git
- Creating a Repository
- Managing versions and tracking changes
- Versioning multiple files
- Intro to GitHub
- Collaborating with GitHub
- BONUS
- Glossary of terms
Background
We’ll start by exploring how version control can be used to keep track of what one person did and when. Even if you aren’t collaborating with other people, automated version control is much better than this situation:
“Piled Higher and Deeper” by Jorge Cham, http://www.phdcomics.com
We’ve all been in this situation before: it seems ridiculous to have multiple nearly-identical versions of the same document. Some word processors let us deal with this a little better, such as Microsoft Word’s Track Changes, Google Docs’ version history, or LibreOffice’s Recording and Displaying Changes.
Version control systems start with a base version of the document and then record changes you make each step of the way. You can think of it as a recording of your progress: you can rewind to start at the base document and play back each change you made, eventually arriving at your more recent version.
Once you think of changes as separate from the document itself, you can then think about “playing back” different sets of changes on the base document, ultimately resulting in different versions of that document. For example, two users can make independent sets of changes on the same document.
Unless multiple users make changes to the same section of the document - a conflict - you can incorporate two sets of changes into the same base document.
A version control system is a tool that keeps track of these changes for us, effectively creating different versions of our files. It allows us to decide which changes will be made to the next version (each record of these changes is called a commit), and keeps useful metadata about them. The complete history of commits for a particular project and their metadata make up a repository. Repositories can be kept in sync across different computers, facilitating collaboration among different people.
Paper Writing
Imagine you drafted an excellent paragraph for a paper you are writing, but later ruin it. How would you retrieve the -excellent- version of your conclusion? Is it even possible?
Imagine you have 5 co-authors. How would you manage the changes and comments they make to your paper? If you use LibreOffice Writer or Microsoft Word, what happens if you accept changes made using the
Track Changesoption? Do you have a history of those changes?Solution
Recovering the excellent version is only possible if you created a copy of the old version of the paper. The danger of losing good versions often leads to the problematic workflow illustrated in the PhD Comics cartoon at the top of this page.
Collaborative writing with traditional word processors is cumbersome. Either every collaborator has to work on a document sequentially (slowing down the process of writing), or you have to send out a version to all collaborators and manually merge their comments into your document. The ‘track changes’ or ‘record changes’ option can highlight changes for you and simplifies merging, but as soon as you accept changes you will lose their history. You will then no longer know who suggested that change, why it was suggested, or when it was merged into the rest of the document. Even online word processors like Google Docs or Microsoft Office Online do not fully resolve these problems.
Setting up Git
When we use Git on a new computer for the first time, we need to configure a few things. Below are a few examples of configurations we will set as we get started with Git:
- our name and email address,
- what our preferred text editor is,
- and that we want to use these settings globally (i.e. for every project).
On a command line, Git commands are written as git verb options,
where verb is what we actually want to do and options is additional optional information which may be needed for the verb. So here is how
Riley sets up their new laptop:
git config --global user.name "Riley Shor"
git config --global user.email "Riley.Shor@fake.email.address"
Please use your own name and email address instead of Riley’s. This user name and email will be associated with your subsequent Git activity, which means that any changes pushed to GitHub, BitBucket, GitLab or another Git host server in a later lesson will include this information.
For these lessons, we will be interacting with GitHub and so the email address used should be the same as the one used when setting up your GitHub account. If you are concerned about privacy, please review GitHub’s instructions for keeping your email address private.
GitHub, GitLab, & BitBucket
GitHub, GitLab, & BitBucket are websites where you can store your git repositories, share them with the world, and collaborate with others. You can think of them like email applications. You may have a gmail address, and you can choose to manage your email through one of many services such as the Gmail app, Microsoft Outlook, Apple’s Mail app, etc. They have different interfaces and features, but all of them allow you to manage your email. Similarly, GitHub, GitLab, & BitBucket have different interfaces and features, but they all allow you to store, share, and collaborate with others on your git repos.
Line Endings
As with other keys, when you hit Return on your keyboard, your computer encodes this input as a character. Different operating systems use different character(s) to represent the end of a line. (You may also hear these referred to as newlines or line breaks.) Because Git uses these characters to compare files, it may cause unexpected issues when editing a file on different machines. Though it is beyond the scope of this lesson, you can read more about this issue in the Pro Git book.
You can change the way Git recognizes and encodes line endings using the
core.autocrlfcommand togit config. The following settings are recommended:On macOS and Linux:
git config --global core.autocrlf inputAnd on Windows:
git config --global core.autocrlf true
This optional setting will make a downstream step with GitHub a bit clearer.
git config --global init.defaultBranch main
Riley also has to set their favorite text editor, nano.
git config --global core.editor "nano -w"
Git can be configured to support most text editors. You can reconfigure the text editor for Git whenever you want to change it.
| Editor | Configuration command |
|---|---|
| Atom | $ git config --global core.editor "atom --wait" |
| nano | $ git config --global core.editor "nano -w" |
| BBEdit (Mac, with command line tools) | $ git config --global core.editor "bbedit -w" |
| Sublime Text (Mac) | $ git config --global core.editor "/Applications/Sublime\ Text.app/Contents/SharedSupport/bin/subl -n -w" |
| Sublime Text (Win, 32-bit install) | $ git config --global core.editor "'c:/program files (x86)/sublime text 3/sublime_text.exe' -w" |
| Sublime Text (Win, 64-bit install) | $ git config --global core.editor "'c:/program files/sublime text 3/sublime_text.exe' -w" |
| Notepad (Win) | $ git config --global core.editor "c:/Windows/System32/notepad.exe" |
| Notepad++ (Win, 32-bit install) | $ git config --global core.editor "'c:/program files (x86)/Notepad++/notepad++.exe' -multiInst -notabbar -nosession -noPlugin" |
| Notepad++ (Win, 64-bit install) | $ git config --global core.editor "'c:/program files/Notepad++/notepad++.exe' -multiInst -notabbar -nosession -noPlugin" |
| Kate (Linux) | $ git config --global core.editor "kate" |
| Gedit (Linux) | $ git config --global core.editor "gedit --wait --new-window" |
| Scratch (Linux) | $ git config --global core.editor "scratch-text-editor" |
| Emacs | $ git config --global core.editor "emacs" |
| Vim | $ git config --global core.editor "vim" |
| VS Code | $ git config --global core.editor "code --wait" |
Vim is the default editor. If did not change your editor and stuck in this editor, the following instructions will help you exit.
Exiting Vim
Note that Vim is the default editor for many programs. If you haven’t used Vim before and wish to exit a session without saving your changes, press Esc then type
:q!and hit Return. If you want to save your changes and quit, press Esc then type:wqand hit Return.
The four commands we just ran above only need to be run once: the flag --global tells Git
to use the settings for every project, in your user account, on this computer.
You can check your global settings at any time:
git config --global --list
You can change your configuration as many times as you want: use the same commands to choose another editor or update your email address.
Proxy
In some networks you need to use a proxy. If this is the case, you may also need to tell Git about the proxy:
git config --global http.proxy proxy-url git config --global https.proxy proxy-urlTo disable the proxy, use
git config --global --unset http.proxy git config --global --unset https.proxy
Git Help and Manual
Always remember that if you forget a
gitcommand, you can access the list of commands by using-hand access the Git manual by using--help:git config -h git config --helpWhile viewing the manual, remember the
:is a prompt waiting for commands and you can press Q to exit the manual.
Creating a Repository
Once Git is configured, we can start using it. We’ll start with a simple example
to explore a few of the most important and common git commands. For this example,
we will create a new project directory called cheatsheets inside our home
directory:
cd # cd by itself changes to the home directory
mkdir cheatsheets
cd cheatsheets
pwd
/home/USERNAME/Desktop/cheatsheets
Let’s create a file called bash-cheatsheet.txt. We’ll write some notes about
a few bash commands we learned so far. We’ll use
nano to edit the file (but you can use whatever text editor you like).
nano bash-cheatsheet.txt
Type the text below into the bash-cheatsheet.txt file:
Bash cheatsheet
SWC-Workshop
Let’s first verify that the file was properly created by running the list command (ls):
ls
bash-cheatsheet.txt
Now we tell Git to make the cheatsheets into a repository
– a place where Git can store versions of our files:
git init
Initialized empty Git repository in /Users/USERNAME/Desktop/cheatsheets/.git/
It is important to note that git init will create a repository that
includes subdirectories and their files—there is no need to create
separate repositories nested within the cheatsheets repository, whether
subdirectories are present from the beginning or added later. Also, note
that the creation of the cheatsheets directory and its initialization as a
repository are completely separate processes.
If we use ls to show the directory’s contents,
it appears that nothing has changed:
ls
But if we add the -a flag to show everything,
we can see that Git has created a hidden directory within cheatsheets called .git:
ls -a -F
./ ../ .git/ bash-cheatsheet.txt
Git uses this special subdirectory to store all the information about the project,
including all files and sub-directories located within the project’s directory.
If we ever delete the .git subdirectory,
we will lose the project’s history.
We can check that everything is set up correctly by asking Git to tell us the status of our project:
git status
On branch main
No commits yet
Untracked files:
(use "git add <file>..." to include in what will be committed)
bash-cheatsheet.txt
nothing added to commit but untracked files present (use "git add" to track)
(If you are using a different version of git, the exact
wording of the output might be slightly different.)
Note that Git lists our file bash-cheatsheet.txt in the Untracked files
section. This is Git’s way of saying that it sees a file in the directory that
is not yet part of the repository. We use a separate command to tell git to add
files to the repository.
git add bash-cheatsheet.txt
Git does output any message, but you can check that the file has been added
as a new file with the git status command.
git status
On branch main
Changes to be committed:
(use "git restore --staged <file>..." to unstage)
new file: bash-cheatsheet.txt
The file is ready or staged, but to actually take the snapshot of this version, we have to execute one last command:
git commit -m'Added Bash cheatsheet'
[main 7f8cd28] Added B ash cheatsheet
1 file changed, 2 insertions(+)
We can see that the repository matches the current contents of the directory like so:
git status
On branch main
nothing to commit, working tree clean
The add commit and status commands are the most common and important commands in Git, so
let’s execute this sequence once more below.
nano bash-cheatsheet.txt
Add the new lines below into the bash-cheatsheet.txt file:
Bash cheatsheet
SWC-Workshop
Commands
pwd : path to working dir
cd : change directory
ls : list directory contents
git status
On branch main
Changes not staged for commit:
(use "git add <file>..." to update what will be committed)
(use "git restore <file>..." to discard changes in working directory)
modified: bash-cheatsheet.txt
no changes added to commit (use "git add" and/or "git commit -a")
Note that Git recognizes the file was changed since the last commit.
We will add and commit these new changes.
git add bash-cheatsheet.txt
git commit -m'Added basic commands'
[main 6441a95] Added email and date
1 file changed, 3 insertions(+)
And again checking status, Git shows that the repository’s current version
matches the directory contents.
git status
On branch main
nothing to commit, working tree clean
You can see from above that Git insists that we add files to the set we want to commit before actually committing anything. This allows us to commit our changes together and capture changes in logical portions rather than only large batches.
For example, suppose we’re adding a few citations to relevant research to our thesis. We might want to commit those additions, and the corresponding bibliography entries, but -not- commit some of our work drafting the conclusion (which we haven’t finished yet).
To allow for this, Git has a special -staging area- where it keeps track of things that have been added to the current changeset but not yet committed.
Staging Area
If you think of Git as taking snapshots of changes over the life of a project,
git addspecifies -what- will go in a snapshot (putting things in the staging area), andgit committhen -actually takes- the snapshot, and makes a permanent record of it (as a commit). If you don’t have anything staged when you typegit commit, Git will prompt you to usegit commit -aorgit commit --all, which is kind of like gathering -everyone- to take a group photo! However, it’s almost always better to explicitly add things to the staging area, because you might commit changes you forgot you made. (Going back to the group photo simile, you might get an extra with incomplete makeup walking on the stage for the picture because you used-a!) Try to stage things manually, or you might find yourself searching for “how to undo a commit” more than you would like! We’ll show you how to do this a little later in this lesson.
Managing Versions and Tracking Changes
So far, we focused on adding files and changes to a repository. We will reinforce those commands in a similar exercise, but also introduce new git commands to view and manage version history of the repository.
We’ll start by creating a new project related to the plots you created earlier.
cd # cd by itself changes to the home directory
mkdir un-report-readme
cd un-report-readme
pwd
/home/USERNAME/Desktop/un-report-readme
What should git status return on a brand new directory?
git status
fatal: not a git repository (or any of the parent directories): .git
So we initialize the repository:
git init
Initialized empty Git repository in /Users/USERNAME/Desktop/un-report-readme/.git/
Let’s create a file called README.md. We’ll write some notes about
some of the plots we created earlier. We’ll use
nano to edit the file (but you can use whatever text editor you like).
nano README.md
Type the text below into the README.md file:
# UN Report
10/18/2021
Riley.Shor@fake.email.address
And add and commit that new file:
git add README.md
git commit -m'Added README'
git status
[main (root-commit) ac0b28a] Added README
1 file changed, 4 insertions(+)
create mode 100644 README.md
On branch main
nothing to commit, working tree clean
For practice, we’ll make a series of changes, adding and committing after each change. For the first change, in your editor, add the summary line shown below.
# UN Report
10/18/2021
Riley.Shor@fake.email.address
Scripts to read and interpret gapminder data.
git add README.md
git commit -m'Added a summary of the project'
[main 4fec7ab] Added a summary of the project
1 file changed, 1 insertions(+)
One more change: revise the summary line as shown below.
# UN Report
10/18/2021
Riley.Shor@fake.email.address
Scripts to read and interpret gapminder data producing figures that emphasize
the relationship of GDP and life expectancy across countries.
git add README.md
git commit -m'Updated summary with details on figures'
[main a6669c4] Added a summary of the project
1 file changed, 1 insertions(+), 1 deletions(-)
git status
On branch main
nothing to commit, working tree clean
Every time you commit a change, Git takes a snapshot of the file. You can see
the full list of snapshots with git log.
git log
commit a6669c4fc2a598677a660ce5b831b1e2c9445a96 (HEAD -> main)
Author: Riley Shor <Riley.Shor@fake.email.address>
Date: Sun Oct 17 10:00:06 2021 -0400
Updated summary with details on figures
commit 4fec7ab0937addcdebf41962f797bcd92fd1295b
Author: Riley Shor <Riley.Shor@fake.email.address>
Date: Sun Oct 17 09:59:16 2021 -0400
Added a summary of the project
commit ac0b28aa38d617eada4a87f383cb7abc51a5faaa
Author: Riley Shor <Riley.Shor@fake.email.address>
Date: Sun Oct 17 09:58:38 2021 -0400
Added README
git log lists all commits made to a repository in reverse chronological order.
The listing for each commit includes:
- the commit’s full identifier (which starts with the same characters as
the short identifier printed by the
git commitcommand earlier), - the commit’s author,
- when it was created,
- and the commit message
Often times, a shorter log format is just as useful and easier to read. The
--oneline flag simply shows an abbreviated identifier and the commit comment.
git log --oneline
a6669c4 (HEAD -> main) Updated summary with details on figures
4fec7ab Added a summary of the project
ac0b28a Added README
Each Git commit tracks the content and circumstances of all the changes. You
can see more info on a specific commit with git show followed by one of the
commit identifiers from log command above:
git show 4fec7ab0937addcdebf41962f797bcd92fd1295b
commit 4fec7ab0937addcdebf41962f797bcd92fd1295b
Author: Riley Shor <Riley.Shor@fake.email.address>
Date: Sun Oct 17 09:59:16 2021 -0400
Added a summary of the project
diff --git a/README.md b/README.md
index 5a6bf77..03d7655 100644
--- a/README.md
+++ b/README.md
@@ -2,3 +2,4 @@
10/18/2021
Riley.Shor@fake.email.address
+Scripts to read and interpret gapminder data.
git show outputs the info from log followed by the differences between the
specified commit and the preceding version. It’s a lot of information, so
we’ll break it down into pieces:
- The first line tells us that Git is producing output similar to the Unix
diffcommand comparing the old and new versions of the file. - The second line tells exactly which versions of the file
Git is comparing;
5a6bf77and03d7655are unique computer-generated labels for those versions. - The third and fourth lines once again show the name of the file being changed.
- The remaining lines are the most interesting, they show us the actual differences
and the lines on which they occur. In particular, it starts with a few
lines in common to give you some context (“10/18/2021” and
“Riley.Shor@fake.email.address”); the final lines show a line was added
to the new version (
+).
Git commit identifiers
Git needs a long commit identifier to ensure all identifiers are unique. That said, most git operations that use a commit identifier can use a shorter identifier provided it’s unambiguous for that repository’s list of existing commits. It is common to use only the first few characters (~7 characters) when specifying a identifier.
Also, we will continue to use the term “commit identifier”, but identifiers are sometimes referred to as “commit index”, “commit hash”, or “commit SHA” (sounds like “shah”).
This command shows the latest commit using an abbreviated commit identifier:
git show a6669c4
commit a6669c4fc2a598677a660ce5b831b1e2c9445a96 (HEAD -> main)
Author: Riley Shor <Riley.Shor@fake.email.address>
Date: Sun Oct 17 10:00:06 2021 -0400
Updated summary with details on figures
diff --git a/README.md b/README.md
index 03d7655..71663b6 100644
--- a/README.md
+++ b/README.md
@@ -2,4 +2,5 @@
10/18/2021
Riley.Shor@fake.email.address
-Scripts to read and interpret gapminder data.
+Scripts to read and interpret gapminder data producing figures that emphasize
+the relationship of GDP and life expectancy across countries.
As detailed in the comment above, the trailing output shows the actual change from the previous version of README.md.
10/18/2021
Riley.Shor@fake.email.address
Then it shows that one line was deleted from the previous version and two lines were added to new version.
-Scripts to read and interpret gapminder data.
+Scripts to read and interpret gapminder data producing figures that emphasize
+the relationship of GDP and life expectancy across countries.
In addition to comparing versions already committed, you can also compare
in-progress changes to previous commits using git diff. To see this in action,
add the following 4 trailing lines to README.md (starting with “Files/dirs:”).
# UN Report
10/18/2021
Riley.Shor@fake.email.address
Scripts to read and interpret gapminder data producing figures that emphasize
the relationship of GDP and life expectancy across countries.
Files/dirs:
code : R scripts
data : gapminder text data
figures : plots
Check that there is a difference between the working dir and the last repo:
git status
On branch main
Changes not staged for commit:
(use "git add <file>..." to update what will be committed)
(use "git restore <file>..." to discard changes in working directory)
modified: README.md
no changes added to commit (use "git add" and/or "git commit -a")
See the actual differences between the working dir of the file and the last
commit with git diff:
git diff README.md
diff --git a/README.md b/README.md
index 71663b6..a5a8954 100644
--- a/README.md
+++ b/README.md
@@ -4,3 +4,8 @@ Riley.Shor@fake.email.address
Scripts to read and interpret gapminder data producing figures that emphasize
the relationship of GDP and life expectancy across countries.
+
+Files/dirs:
+code : R scripts
+data : gapminder text data
+figures : plots
Sometimes you realize you want to undo a change after you have added the file.
git restore accomplishes this. To illustrate this pattern, we’ll add this
change to the staging area:
git add README.md
git status
On branch main
Changes to be committed:
(use "git restore --staged <file>..." to unstage)
modified: README.md
The git restore --staged command “unstages” the change, keeping the changes in
the working file.
git restore --staged README.md
cat README.md
# UN Report
10/18/2021
Riley.Shor@fake.email.address
Scripts to read and interpret gapminder data producing figures that emphasize
the relationship of GDP and life expectancy across countries.
Files/dirs:
code : R scripts
data : gapminder text data
figures : plots
git status
On branch main
Changes not staged for commit:
(use "git add <file>..." to update what will be committed)
(use "git restore <file>..." to discard changes in working directory)
modified: README.md
no changes added to commit (use "git add" and/or "git commit -a")
You can use git restore --staged to abandon the working changes and replace with the
version from the last commit:
git restore --staged README.md
git status
At times, it might make sense to abandon the changes in the working dir and replace with the last commit.
git restore README.md
git status
On branch main
nothing to commit, working tree clean
Consider the git restore command in the context of the earlier diagram:
git checkout allows you to peek at an earlier version of the project without
making any changes. Before you do this, verify you have no outstanding
modifications. Use some combination of restore or add and commit so that
you have a clean working tree.
git status
git log --oneline
On branch main
nothing to commit, working tree clean
a6669c4 (HEAD -> main) Updated summary with details on figures
4fec7ab Added a summary of the project
ac0b28a Added README
Checkout the initial commit (will be the last in your list).
git checkout ac0b28a
Note: switching to 'ac0b28a'.
You are in 'detached HEAD' state. You can look around, make experimental
changes and commit them, and you can discard any commits you make in this
state without impacting any branches by switching back to a branch.
If you want to create a new branch to retain commits you create, you may
do so (now or later) by using -c with the switch command. Example:
git switch -c <new-branch-name>
Or undo this operation with:
git switch -
Turn off this advice by setting config variable advice.detachedHead to false
HEAD is now at ac0b28a Added README
The “detached HEAD” is like “look, but don’t touch” here, so you shouldn’t make any changes in this state.
Git has rewound the clock and dropped you into an earlier version. Note the log shows only the one change and git has replaced your working dir with the earliest version of this file.
git log --oneline
cat README
ac0b28a (HEAD) Added README
# UN Report
10/18/2021
Riley.Shor@fake.email.address
When you are done looking, use git checkout main to return to the latest
version.
git checkout main
git log --oneline
Previous HEAD position was ac0b28a Added README
Switched to branch 'main'
a6669c4 (HEAD -> main) Updated summary with details on figures
4fec7ab Added a summary of the project
ac0b28a Added README
Finally, if you are having second thoughts about a change you have already
commited, you can revert the change. To illustrate this, we will delete
our README.md file, commit the change, and then revert the change.
rm README.md
git status
On branch main
Changes not staged for commit:
(use "git add/rm <file>..." to update what will be committed)
(use "git restore <file>..." to discard changes in working directory)
deleted: README.md
no changes added to commit (use "git add" and/or "git commit -a")
Note that Git recognizes the file was deleted. We still use
git addto add the change to the staging area.
git add README.md
git status
On branch main
Changes to be committed:
(use "git restore --staged <file>..." to unstage)
deleted: README.md
git commit -m'Deleted README'
[main 24c3a76] Deleted README
1 file changed, 6 deletions(-)
delete mode 100644 README.md
(base)
The file is not in the working directory and the Git log shows the commit that deleted it.
ls
git log --oneline
24c3a76 (HEAD -> main) Deleted README
a6669c4 Updated summary with details on figures
4fec7ab Added a summary of the project
ac0b28a Added README
So now we revert the last commit to rescue that file. This command will launch an editor to allow you to edit a new commit message. You can accept the default message and save/exit the editor.
git revert 24c3a76
[main 4a82b63] Revert "Deleted README"
1 file changed, 6 insertions(+)
create mode 100644 README.md
Note that the file was restored to its previous state and Git has added a new commit. This shows that when Git reverts a change, it does not “go back in time” to before the problem commit. Instead it creates a new change that inverts the effect of the problem commit; in our case recreating the file.
git log --oneline cat README.md
4a82b63 (HEAD -> main) Revert "Deleted README"
24c3a76 Deleted README
a6669c4 Updated summary with details on figures
4fec7ab Added a summary of the project
ac0b28a Added README
# UN Report
10/18/2021
Riley.Shor@fake.email.address
Scripts to read and interpret gapminder data producing figures that emphasize
the relationship of GDP and life expectancy across countries.
Versioning multiple files
Up until now, we have used git with a single file. Git’s real power is that you can use these same commands to version a set of files all at once, enabling coordinated changes that keep all the files in a consistent state. We will explore this by versioning un-report dir we were working in earlier.
cd
cd un-report
pwd
/home/USERNAME/Desktop/cheatsheets
Initialize a new Git repository.
git init
git status
Initialized empty Git repository in /Users/USERNAME/Desktop/un-report/.git/
On branch main
No commits yet
Untracked files:
(use "git add <file>..." to include in what will be committed)
.Rproj.user/
code/
data/
figures/
un-report.Rproj
nothing added to commit but untracked files present (use "git add" to track)
We can add directories in the same we added a single file.
git add code
git status
On branch main
No commits yet
Changes to be committed:
(use "git rm --cached <file>..." to unstage)
new file: code/gdp_population.R
Untracked files:
(use "git add <file>..." to include in what will be committed)
.Rproj.user/
data/
figures/
un-report.Rproj
Note that when adding the directory, Git assumes you want to add everything inside that directory. For our purposes that’s correct. Let’s continue by adding the
dataandfiguresdirectories. We can do this in one command:
git add data figures
git status
On branch main
No commits yet
Changes to be committed:
(use "git rm --cached <file>..." to unstage)
new file: code/gdp_population.R
new file: data/co2-un-data.csv
new file: data/gapminder_1997.csv
new file: data/gapminder_data.csv
new file: data/rnd-un-data.csv
new file: figures/awesome_plot.jpg
new file: figures/awesome_violin_plot.jpg
Untracked files:
(use "git add <file>..." to include in what will be committed)
.Rproj.user/
un-report.Rproj
We could keep adding files, but let’s commit what we have now.
git commit -m'Added code, data, figures dirs'
git status
[main (root-commit) 42e0844] Added code, data, figures dirs
7 files changed, 6404 insertions(+)
create mode 100644 code/gdp_population.R
create mode 100644 data/co2-un-data.csv
create mode 100644 data/gapminder_1997.csv
create mode 100644 data/gapminder_data.csv
create mode 100644 data/rnd-un-data.csv
create mode 100644 figures/awesome_plot.jpg
create mode 100644 figures/awesome_violin_plot.jpg
On branch main
Untracked files:
(use "git add <file>..." to include in what will be committed)
.Rproj.user/
un-report.Rproj
nothing added to commit but untracked files present (use "git add" to track)
One of the reasons Git separates the action of adding files to the staging area
and committing the changes to the repository is that it gives allows you to group
commit into a meaningful, logical, consistent set of changes. It also gives you
the freedom to decide that some files should not be committed to the repository.
Moreover, Git can use a .gitignore file to emphasize to Git that it should
leave certain files in the working directory and never try to add them to the
repository.
Choosing what include in a repository and what to leave out
Some kinds of files belong in a Git repository. Some kinds should never be added to Git. And sometimes it depends on the context and the file. In the UN Report example above, there are files for data, code, figures, and also R-Studio configuration. Which of these do you think would be most appropriate to use for version control? Can you think of examples of files that you might not want to add to Git?
Solution
We will discuss this together in the workshop session.
For now, we will choose to add un-report.Rproj to the repository and ignore
the .Rproj.user/ directory. In your text editor, create a new file named
.gitignore and add the following line:
.Rproj.user/
git status now considers the .gitignore file and correctly ignores that dir
(suppressing it in the output).
git status
On branch main
Untracked files:
(use "git add <file>..." to include in what will be committed)
.gitignore
un-report.Rproj
nothing added to commit but untracked files present (use "git add" to track)
Finally, we’ll commit those last changes.
git add un-report.Rproj .gitignore
git commit -m'Added .gitignore and R-Studio project info'
[main 25147ba] Added .gitignore and R-Studio project info
2 files changed, 15 insertions(+)
create mode 100644 .gitignore
create mode 100644 un-report.Rproj
git status
On branch main
nothing to commit, working tree clean
Consider your commit message
Not all commit messages are created equal. Messages can be too short, too long, redundant, or vague. Indeed some of the commit messages i the examples above may be suboptimal. Consider where the commit messages are used and by whom; discuss some characteristics of an ideal commit message.
Solution
We will discuss this together in the workshop session.
A last thought about directories: Git does not track directories on their own, only the files within them.
mkdir project_background git status git add project_background git statusNote, our newly created empty directory
analysisdoes not appear in the list of untracked files even if we explicitly add it (viagit add) to our repository. This is the reason why you will sometimes see.gitkeepfiles in otherwise empty directories. Unlike.gitignore, these files are not special and their sole purpose is to populate a directory so that Git adds it to the repository. In fact, you can name such files anything you like.touch project_background/.gitkeep git status git add project_background git status
Git concepts and commands (so far)
- config
- init
- status
- add
- commit
- restore
- log
- diff
- show
- checkout
- revert
- .gitignore
Intro to GitHub
Now that you’ve created a git repo and gotten the hang of the basic git workflow, it’s time to share your repo with the world. Systems like Git allow us to move work between any two repositories. In practice, though, it’s easiest to use one copy as a central hub, and to keep it on the web rather than on someone’s laptop. Most programmers use hosting services like GitHub, Bitbucket or GitLab to hold those master copies.
Let’s start by sharing the changes we’ve made to our current project with the world. We do this by creating a repository on GitHub and linking it to our existing local repository. To link these, GitHub security requires you to use either a Personal Access Token (like a limited-use password) or an SSH key (which establishes a trusted relationship between two systems). Either way works well; we will use the SSH approach detailed below.
SSH Background and Setup
SSH (Secure Shell Protocol) key is commonly used by many different services to authenticate access on the command line. SSH is a cryptographic network protocol that allows secure communication between computers using an otherwise insecure network.
SSH uses what is called a key pair; two keys that work together to validate access. One key is publicly known (the public key), is kept private (the private key).
You can think of the public key as a padlock, and only you have the key (the private key) to open it. You use the public key where you want a secure method of communication, such as your GitHub account. You give this padlock, or public key, to GitHub and say “lock the communications to my account with this so that only computers that have my private key can unlock communications and send git commands as my GitHub account.”
What we will do now is the minimum required to set up the SSH keys and add the public key to a GitHub account.
Advanced SSH
See GitHub SSH for more details on using and configuring SSH.
The first thing we are going to do is check if this has already been done on the computer you’re on. Because generally speaking, this setup only needs to happen once and then you can forget about it.
First we execute this command to check that no SSH key is already set up in GitHub.
ssh -T git@github.com
The first time you do this, you might see the following. When prompted answer “yes”.
The authenticity of host 'github.com (192.30.255.112)' can't be established.
RSA key fingerprint is SHA256:nThbg6kXUpJWGl7E1IGOCspRomTxdCARLviKw6E5SY8.
This key is not known by any other names
Are you sure you want to continue connecting (yes/no/[fingerprint])? y
Please type 'yes', 'no' or the fingerprint: yes
Warning: Permanently added 'github.com' (RSA) to the list of known hosts.
The next line could one of the following outputs below.
Hi Riley! You've successfully authenticated, but GitHub does not provide shell access.
If you see this, then you SSH access is already set up, so you can proceed to Making a repository on GitHub and linking it to a local repository
git@github.com: Permission denied (publickey).
If you see this, it’s ok; keep going below to set up SSH access.
Keeping your keys secure
You shouldn’t really forget about your SSH keys, since they keep your account secure. It’s good practice to audit your secure shell keys every so often. Especially if you are using multiple computers to access your account.
We will run the list command to check what key pairs already exist on your computer.
ls -al ~/.ssh
Your output is going to look a little different depending on whether or not SSH has ever been set up on the computer you are using.
Riley has not set up SSH on her computer, so her output is
ls: cannot access '/c/Users/Riley/.ssh': No such file or directory
If SSH has been set up on the computer you’re using, the public and private key pairs will be listed. The file names are either id_ed25519/id_ed25519.pub or id_rsa/id_rsa.pub depending on how the key pairs were set up.
Since they don’t exist on Riley’s computer, she uses this command to create them:
ssh-keygen -t ed25519 -C "Riley.Shor@fake.email.address"
If you are using a legacy system that doesn’t support the Ed25519 algorithm, use:
ssh-keygen -t rsa -b 4096 -C "Riley.Shor@fake.email.address"
Generating public/private ed25519 key pair.
Enter file in which to save the key (/c/Users/Riley/.ssh/id_ed25519):
We want to use the default file, so just press Enter.
Created directory '/c/Users/Riley/.ssh'.
Enter passphrase (empty for no passphrase):
Now, it is prompting Riley for a passphrase. Since she is using her lab’s laptop that other people sometimes have access to, she wants to create a passphrase. Be sure to use something memorable or save your passphrase somewhere, as there is no “reset my password” option.
Enter same passphrase again:
After entering the same passphrase a second time, we receive the confirmation
Your identification has been saved in /c/Users/Riley/.ssh/id_ed25519
Your public key has been saved in /c/Users/Riley/.ssh/id_ed25519.pub
The key fingerprint is:
SHA256:SMSPIStNyA00KPxuYu94KpZgRAYjgt9g4BA4kFy3g1o Riley.Shor@fake.email.address
The key's randomart image is:
+--[ED25519 256]--+
|^B== o. |
|%-=.-.+ |
|+=.E =.+ |
| .=.+.o.. |
|.... . S |
|.+ o |
|+ = |
|.o.o |
|oo+. |
+----[SHA256]-----+
The “identification” is actually the private key. You should never share it. The public key is appropriately named. The “key fingerprint” is a shorter version of a public key.
Now that we have generated the SSH keys, we will find the SSH files when we check.
ls -al ~/.ssh
drwxr-xr-x 1 Riley Shor 197121 0 Jul 16 14:48 ./
drwxr-xr-x 1 Riley Shor 197121 0 Jul 16 14:48 ../
-rw-r--r-- 1 Riley Shor 197121 419 Jul 16 14:48 id_ed25519
-rw-r--r-- 1 Riley Shor 197121 106 Jul 16 14:48 id_ed25519.pub
Now we that we have a public and private key pair, we need to give GitHub our public key.
First, we need to copy the public key. Be sure to include the .pub at the end, otherwise you’re looking at the private key.
cat ~/.ssh/id_ed25519.pub
ssh-ed25519 AAAAC3NzaC1lZDI1NTE5AAAAIDmRA3d51X0uu9wXek559gfn6UFNF69yZjChyBIU2qKI Riley.Shor@fake.email.address
Now, going to GitHub.com, click on your profile icon in the top right corner to get the drop-down menu. Click “Settings,” then on the settings page, click “SSH and GPG keys,” on the left side “Account settings” menu. Click the “New SSH key” button on the right side. Now, you can add the title (Riley uses the title “Riley’s Lab Laptop” so she can remember where the original key pair files are located), paste your SSH key into the field, and click the “Add SSH key” to complete the setup.
Now that we’ve set that up, let’s check our authentication again from the command line.
ssh -T git@github.com
Hi Riley! You've successfully authenticated, but GitHub does not provide shell access.
Congratulations: your SSH key is configured correctly. You are ready to begin using GitHub.
Making a repository on GitHub and linking it to a local repository
Log in to GitHub, then click on the icon in the top right corner to
create a new repository called un-report.
Name your repository un-report and then click Create Repository.
Important options
Since this repository will be connected to a local repository, it needs to be empty. Leave “Initialize this repository with a README” unchecked, and keep “None” as options for both “Add .gitignore” and “Add a license.” See the “GitHub License and README files” exercise below for a full explanation of why the repository needs to be empty.
In the screenshots below, the Owner is ‘mkuzak’ and the Repository name is ‘planets’.
You should instead see your own username for the Owner and you should name the
repository un-report.
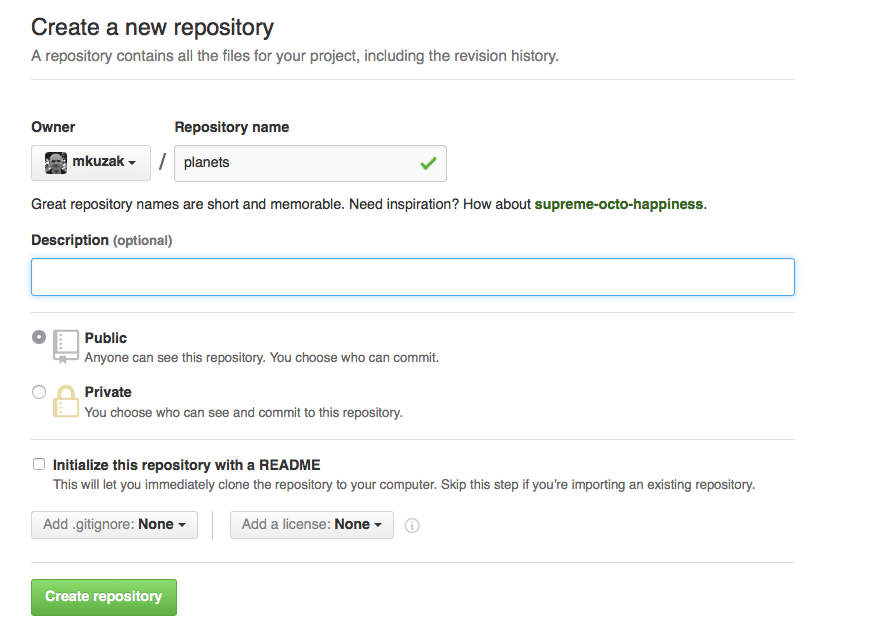
As soon as the repository is created, GitHub displays a page with a URL and some
information on how to configure your local repository. Examine the toggle button
labeled HTTPS | SSH; make sure to select the SSH button.
This effectively does the following on GitHub’s servers:
mkdir un-report
cd un-report
git init
If you remember back to the earlier lesson where we added and
committed our earlier work on un-report, we had a diagram of the local repository
which looked like this:
Now that we have two repositories, we need a diagram like this:
Note that our local repository still contains our earlier work on un-report, but the
remote repository on GitHub appears empty as it doesn’t contain any files yet.
The next step is to connect the two repositories. We do this by making the GitHub repository a remote for the local repository. The home page of the repository on GitHub includes the string we need to identify it:
Copy that URL from the browser, go into the local un-report repository, and run
this command:
git remote add origin git@github.com:USERNAME/un-report.git
Make sure to replace USERNAME with your actual GitHub username so it will use
the correct URL for your repository; that should be the only difference.
origin is a local name used to refer to the remote repository. It could be called
anything, but origin is a convention that is often used by default in git
and GitHub, so it’s helpful to stick with this unless there’s a reason not to.
We can check that the command has worked by running git remote -v:
git remote -v
origin git@github.com:USERNAME/un-report.git (push)
origin git@github.com:USERNAME/un-report.git (fetch)
Once the remote is set up, this command will push the changes from our local repository to the repository on GitHub:
git push -u origin main
Enumerating objects: 16, done.
Counting objects: 100% (16/16), done.
Delta compression using up to 8 threads.
Compressing objects: 100% (11/11), done.
Writing objects: 100% (16/16), 1.45 KiB | 372.00 KiB/s, done.
Total 16 (delta 2), reused 0 (delta 0)
remote: Resolving deltas: 100% (2/2), done.
To git@github.com:USERNAME/un-report.git
- [new branch] main -> main
Our local and remote repositories are now in this state:
The ‘-u’ Flag
You may see a
-uoption used withgit pushin some documentation. This option is synonymous with the--set-upstream-tooption for thegit branchcommand, and is used to associate the current branch with a remote branch so that thegit pullcommand can be used without any arguments. To do this, simply usegit push -u origin mainonce the remote has been set up.
We can pull changes from the remote repository to the local one as well:
git pull origin main
From git@github.com:USERNAME/un-report
- branch main -> FETCH_HEAD
Already up-to-date.
Pulling has no effect in this case because the two repositories are already synchronized. If someone else had pushed some changes to the repository on GitHub, though, this command would download them to our local repository.
GitHub GUI
Browse to your
un-reportrepository on GitHub. Under the Code tab, find and click on the text that says “XX commits” (where “XX” is some number). Hover over, and click on, the three buttons to the right of each commit. What information can you gather/explore from these buttons? How would you get that same information in the shell?Solution
The left-most button (with the picture of a clipboard) copies the full identifier of the commit to the clipboard. In the shell,
git logwill show you the full commit identifier for each commit.When you click on the middle button, you’ll see all of the changes that were made in that particular commit. Green shaded lines indicate additions and red ones removals. In the shell we can do the same thing with
git diff. In particular,git diff ID1..ID2where ID1 and ID2 are commit identifiers (e.g.git diff a3bf1e5..041e637) will show the differences between those two commits.The right-most button lets you view all of the files in the repository at the time of that commit. To do this in the shell, we’d need to checkout the repository at that particular time. We can do this with
git checkout IDwhere ID is the identifier of the commit we want to look at. If we do this, we need to remember to put the repository back to the right state afterwards!
Uploading files directly in GitHub browser
Github also allows you to skip the command line and upload files directly to your repository without having to leave the browser. There are two options. First you can click the “Upload files” button in the toolbar at the top of the file tree. Or, you can drag and drop files from your desktop onto the file tree. You can read more about this on this GitHub page
Push vs. Commit
In this lesson, we introduced the “git push” command. How is “git push” different from “git commit”?
Solution
When we push changes, we’re interacting with a remote repository to update it with the changes we’ve made locally (often this corresponds to sharing the changes we’ve made with others). Commit only updates your local repository.
GitHub License and README files
In this section we learned about creating a remote repository on GitHub, but when you initialized your GitHub repo, you didn’t add a readme or a license file. If you had, what do you think would have happened when you tried to link your local and remote repositories?
Solution
In this case, we’d see a merge conflict due to unrelated histories. When GitHub creates a readme file, it performs a commit in the remote repository. When you try to pull the remote repository to your local repository, Git detects that they have histories that do not share a common origin and refuses to merge.
git pull origin mainwarning: no common commits remote: Enumerating objects: 3, done. remote: Counting objects: 100% (3/3), done. remote: Total 3 (delta 0), reused 0 (delta 0), pack-reused 0 Unpacking objects: 100% (3/3), done. From git@github.com:USERNAME/un-report - branch main -> FETCH_HEAD - [new branch] main -> origin/main fatal: refusing to merge unrelated historiesYou can force git to merge the two repositories with the option
--allow-unrelated-histories. Be careful when you use this option and carefully examine the contents of local and remote repositories before merging.git pull --allow-unrelated-histories origin mainFrom git@github.com:USERNAME/un-report - branch main -> FETCH_HEAD Merge made by the 'recursive' strategy. README.md | 1 + 1 file changed, 1 insertion(+) create mode 100644 README.md
Collaborating with GitHub
For the next step, get into pairs. One person will be the “Owner” and the other will be the “Collaborator”. The goal is that the Collaborator add changes into the Owner’s repository. We will switch roles at the end, so both persons will play Owner and Collaborator.
Practicing By Yourself
If you’re working through this lesson on your own, you can carry on by opening a second terminal window. This window will represent your partner, working on another computer. You won’t need to give anyone access on GitHub, because both ‘partners’ are you.
The Owner needs to give the Collaborator access. On GitHub, click the settings button on the right, select Manage access, click Invite a collaborator, and then enter your partner’s username.
To accept access to the Owner’s repo, the Collaborator needs to go to https://github.com/notifications. Once there she can accept access to the Owner’s repo.
Next, the Collaborator needs to download a copy of the Owner’s repository to her
machine. This is called “cloning a repo”. To clone the Owner’s repo into
her Desktop folder, the Collaborator enters:
git clone git@github.com:USERNAME/un-report.git ~/Desktop/USERNAME-un-report
Replace USERNAME with the Owner’s username.
The Collaborator can now make a change in her clone of the Owner’s repository, exactly the same way as we’ve been doing before:
cd ~/Desktop/USERNAME-un-report
nano README.md
cat README.md
You can write anything you like. Now might be a good time to list the –dependencies– of the project – the tools and packages that are needed to run the code.
Dependencies:
- R >= 4.0
- tidyverse
git add README.md
git commit -m "List dependencies"
1 file changed, 1 insertion(+)
create mode 100644 README.md
Then push the change to the -Owner’s repository- on GitHub:
git push origin main
Enumerating objects: 4, done.
Counting objects: 4, done.
Delta compression using up to 4 threads.
Compressing objects: 100% (2/2), done.
Writing objects: 100% (3/3), 306 bytes, done.
Total 3 (delta 0), reused 0 (delta 0)
To git@github.com:USERNAME/un-report.git
9272da5..29aba7c main -> main
Note that we didn’t have to create a remote called origin: Git uses this
name by default when we clone a repository. (This is why origin was a
sensible choice earlier when we were setting up remotes by hand.)
Take a look at the Owner’s repository on its GitHub website now (you may need to refresh your browser.) You should be able to see the new commit made by the Collaborator.
To download the Collaborator’s changes from GitHub, the Owner now enters:
git pull origin main
remote: Enumerating objects: 4, done.
remote: Counting objects: 100% (4/4), done.
remote: Compressing objects: 100% (2/2), done.
remote: Total 3 (delta 0), reused 3 (delta 0), pack-reused 0
Unpacking objects: 100% (3/3), done.
From git@github.com:USERNAME/un-report
- branch main -> FETCH_HEAD
9272da5..29aba7c main -> origin/main
Updating 9272da5..29aba7c
Fast-forward
README.md | 1 +
1 file changed, 1 insertion(+)
create mode 100644 README.md
Now the three repositories (Owner’s local, Collaborator’s local, and Owner’s on GitHub) are back in sync!
A Basic Collaborative Workflow
In practice, it is good to be sure that you have an updated version of the repository you are collaborating on, so you should
git pullbefore making your changes. The basic collaborative workflow would be:
- update your local repo with
git pull,- make your changes and stage them with
git add,- commit your changes with
git commit -m, and- upload the changes to GitHub with
git pushIt is better to make many commits with smaller changes rather than one commit with massive changes: small commits are easier to read and review.
Switch Roles and Repeat
Switch roles and repeat the whole process.
Review Changes
The Owner pushed commits to the repository without giving any information to the Collaborator. How can the Collaborator find out what has changed on GitHub?
Solution
On GitHub, the Collaborator can go to the repository and click on “commits” to view the most recent commits pushed to the repository.
Comment Changes in GitHub
The Collaborator has some questions about one line change made by the Owner and has some suggestions to propose.
With GitHub, it is possible to comment the diff of a commit. Over the line of code to comment, a blue comment icon appears to open a comment window.
The Collaborator posts comments and suggestions using GitHub interface.
Version History, Backup, and Version Control
Some backup software (e.g. Time Machine on macOS, Google Drive) can keep a history of the versions of your files. They also allow you to recover specific versions. How is this functionality different from version control? What are some of the benefits of using version control, Git and GitHub?
Solution
Automated backup software gives you less control over how often backups are created and it is often difficult to compare changes between backups. However, Git has a steeper learning curve than backup software. Advantages of using Git and GitHub for version control include:
- Great control over which files to include in commits and when to make commits.
- Very popular way to collaborate on code and analysis projects among programmers, data scientists, and researchers.
- Free and open source.
- GitHub allows you to share your project with the world and accept contributions from outside collaborators.
Some more about remotes
In this episode and the previous one, our local repository has had a single “remote”, called
origin. A remote is a copy of the repository that is hosted somewhere else, that we can push to and pull from, and there’s no reason that you have to work with only one. For example, on some large projects you might have your own copy in your own GitHub account (you’d probably call thisorigin) and also the main “upstream” project repository (let’s call thisupstreamfor the sake of examples). You would pull fromupstreamfrom time to time to get the latest updates that other people have committed.Remember that the name you give to a remote only exists locally. It’s an alias that you choose - whether
origin, orupstream, orfred- and not something intrinstic to the remote repository.The
git remotefamily of commands is used to set up and alter the remotes associated with a repository. Here are some of the most useful ones:
git remote -vlists all the remotes that are configured (we already used this in the last episode)git remote add [name] [url]is used to add a new remotegit remote remove [name]removes a remote. Note that it doesn’t affect the remote repository at all - it just removes the link to it from the local repo.git remote set-url [name] [newurl]changes the URL that is associated with the remote. This is useful if it has moved, e.g. to a different GitHub account, or from GitHub to a different hosting service. Or, if we made a typo when adding it!git remote rename [oldname] [newname]changes the local alias by which a remote is known - its name. For example, one could use this to changeupstreamtofred.
Glossary
Key concepts
- .git/ : hidden dir at the top of the project that contains all of gits internals about the repository; you typically don’t interact with the contents of this directory directly but instead use git commands above.
- staging area : the set of changes files that are ready to be committed
- commit: a version of the project; each version will be assigned a unique commit index
- .gitignore: a way to exclude files from the git repo based on their name
- SSH key: establishes a secure trust relationship between two systems (e.g. your laptop and GitHub)
- GitHub: a website that hosts Git repositories for sharing and collaboration.
Key commands:
- commands to interact with local repository:
- config : configure an option (like user.email); can be global or local to a repository.
- init : initialize a dir as a Git repository.
- add : mark a file should be included in the next commit.
- commit : create a new version of the project using the files in the staging area
- status : list the state of the working dir vs staging area vs last commit.
- restore : abandon changes in progress and switch to the last committed version.
- log : show versions (commits) from newest to oldest changes.
- diff: show the differences between one version and another
- show: combines output of log and diff above for a single commit
- checkout: wind the clock back and have a look at an earlier commit
- revert: undo a commit; more precisely add a new commit that inverts the change in a previous commit
- commands to interact with remote repository
- remote : list or add a remote repository
- clone : initialize a local repository based on a remote
- push : move local changes into remote repository
- pull : move remote changes into local repository
Key Points
Version control is like an unlimited ‘undo’.
Version control also allows many people to work in parallel.
R for Data Analysis
Overview
Teaching: 150 min
Exercises: 15 minQuestions
How can I summarize my data in R?
How can R help make my research more reproducible?
How can I combine two datasets from different sources?
How can data tidying facilitate answering analysis questions?
Objectives
To become familiar with the functions of the
dplyrpackage.To be able to use
dplyrto prepare data for analysis.To be able to combine two different data sources using joins.
To be able to create plots and summary tables to answer analysis questions.
Contents
- Getting started
- An introduction to data analysis in R using
dplyr - Cleaning up data
- Joining data frames
- Analyzing combined data
- Putting it all together
Getting Started
First, navigate to the un-reports directory however you’d like and open un-report.Rproj.
This should open the un-report R project in RStudio.
You can checkt his by seeing if the Files in the bottom right of RStudio are the ones in your un-report directory.
Yesterday we spent a lot of time making plots in R using the ggplot2 package. Visualizing data using plots is a very powerful skill in R, but what if we would like to work with only a subset of our data? Or clean up messy data, calculate summary statistics, create a new variable, or join two datasets together? There are several different methods for doing this in R, and we will touch on a few today using functions the dplyr package.
First, we will create a new RScript file for our work. Open RStudio. Choose “File” > “New File” > “RScript”. We will save this file as un_data_analysis.R
Loading in the data
We will start by importing the complete gapminder dataset that we used yesterday into our fresh new R session. Yesterday we did this using a “point-and-click” commands. Today let’s type them into the console ourselves: gapminder_data <- read_csv("data/gapminder_data.csv")
Exercise
If we look in the console now, we’ll see we’ve received an error message saying that R “could not find the function
read_csv()”. Hint: Packages…Solution
What this means is that R cannot find the function we are trying to call. The reason for this usually is that we are trying to run a function from a package that we have not yet loaded. This is a very common error message that you will probably see a lot when using R. It’s important to remember that you will need to load any packages you want to use into R each time you start a new session. The
read_csvfunction comes from thereadrpackage which is included in thetidyversepackage so we will just load thetidyversepackage and run the import code again.
Now that we know what’s wrong, We will use the read_csv() function from the Tidyverse readr package. Load the tidyverse package and gapminder dataset using the code below.
library(tidyverse)
── Attaching packages ───────────────────────────────────────────────────────────────────────────────────────────────────────────────────── tidyverse 1.3.1 ──
✔ ggplot2 3.3.5 ✔ purrr 0.3.4
✔ tibble 3.1.5 ✔ dplyr 1.0.7
✔ tidyr 1.1.4 ✔ stringr 1.4.0
✔ readr 2.0.2 ✔ forcats 0.5.1
── Conflicts ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
The output in your console shows that by doing this, we attach several useful packages for data wrangling, including readr. Check out these packages and their documentation at tidyverse.org
Reminder: Many of these packages, including
dplyr, come with “Cheatsheets” found under the Help RStudio menu tab.
Reload your data:
gapminder_data <- read_csv("data/gapminder_data.csv")
Rows: 1704 Columns: 6
── Column specification ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
Delimiter: ","
chr (2): country, continent
dbl (4): year, pop, lifeExp, gdpPercap
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
Notice that the output of the read_csv() function is pretty informative. It tells us the name of all of our column headers as how it interpreted the data type. This birds-eye-view can help you take a quick look that everything is how we expect it to be.
Now we have the tools necessary to work through this lesson.
An introduction to data analysis in R using dplyr
Get stats fast with summarize()
Let’s say we would like to know what is the mean (average) life expecteny in the dataset. R has a built in function function called mean() that will calculate this value for us. We can apply that function to our lifeExp column using the summarize() function. Here’s what that looks like:
summarize(gapminder_data, averageLifeExp=mean(lifeExp))
# A tibble: 1 × 1
averageLifeExp
<dbl>
1 59.5
When we call summarize(), we can use any of the column names of our data object as values to pass to other functions. summarize() will return a new data object and our value will be returned as a column.
Note: The
summarize()andsummarise()perform identical functions.
We name this new column so we can use in a future argument. So the averageLifeExp= part tells summarize() to use “averageLifeExp” as the name of the new column. Note that you don’t have to quotes around this new name as long as it starts with a letter and doesn’t include a space.
Instead of including the data as an argument, we can use the pipe operator %>% to pass the data value into the summarize function.
gapminder_data %>% summarize(averageLifeExp=mean(lifeExp))
# A tibble: 1 × 1
averageLifeExp
<dbl>
1 59.5
This line of code will do the exact same thing as our first summary command, but the piping function tells R to use the gapminder_data dataframe as the first argument in the next function.
This lets us “chain” together multiple functions, which will be helpful later.
We can also add an
gapminder_data %>%
summarize(averageLifeExp=mean(lifeExp))
# A tibble: 1 × 1
averageLifeExp
<dbl>
1 59.5
Using the pipe operator %>% and enter command makes our code more readable. The pipe operator %>% also helps to avoid using nested function and minimizes the need for new variables.
Since we use the pipe operator so often, there is a keyboard shortcut for it in RStudio. You can press
Helpful hint: Saving a new dataframe Notice that when we run the following code, we are not actually saving a new variable:
gapminder_data %>% summarize(averageLifeExp=mean(lifeExp))This simply outputs what we have created, but does not change actually change
gapminder_dataor save a new dataframe. To save a new dataframe, we could run:gapminder_data_summarized <- gapminder_data %>% summarize(averageLifeExp=mean(lifeExp))Or if we want to change
gapminder_dataitself:gapminder_data <- gapminder_data %>% summarize(averageLifeExp=mean(lifeExp))IMPORTANT: This would overwrite the existing
gapminder_dataobject.For now, we will not be saving dataframes, since we are just experimenting with
dyplrfunctions, but it will be useful later on in this lesson.
Narrow down rows with filter()
Let’s take a look at the value we just calculated, which tells us the average life expectancy for all rows in the data was 59.5. That seems a bit low, doesn’t it? What’s going on?
Well, remember the dataset contains rows from many different years and many different countries. It’s likely that life expectancy has increased overtime, so it may not make sense to average over all the years at the same time.
Use summarize() to find the most recent year in the data set. We can use the max() function to return the maximum value.
Exercise: Practice using the
%>%to summarize dataFind the mean population using the piping function.
Solution:
gapminder_data %>% summarize(recent_year = max(year))# A tibble: 1 × 1 recent_year <dbl> 1 2007
So we see that the most recent year in the dataset is 2007. Let’s calculate the life expectancy for all countries for only that year. To do that, we will use the filter() function to only use rows for that year before calculating the mean value.
gapminder_data %>%
filter(year == 2007) %>%
summarize(average=mean(lifeExp))
# A tibble: 1 × 1
average
<dbl>
1 67.0
Exercise: Filtering the dataset
What is the average GDP per capita for the first year in the dataset? Hint: the column headers identified by
read_csv()showed us there was a column called gdpPercap in the datasetSolution
Identify the earliest year in our dataset using
min()andsummarize()gapminder_data %>% summarize(first_year=min(year))# A tibble: 1 × 1 first_year <dbl> 1 1952We see here that the first year in the dataset is 1952. Filter to only 1952, and determin the average GDP per capita.
gapminder_data %>% filter(year == 1952) %>% summarize(average_gdp=mean(gdpPercap))# A tibble: 1 × 1 average_gdp <dbl> 1 3725.By combining
filter()andsummarize()we were able to calculate the mean GDP per capita in the year 1952.
Notice how the pipe operator (%>%) allows us to combine these two simple steps into a more complicated data extraction?. We took the data, filtered out the rows, then took the mean value. The argument we pass to filter() needs to be some expression that will return TRUE or FALSE. We can use comparisons like > (greater than) and < (less than) for example. Here we tested for equality using a double equals sign ==. You use == (double equals) when testing if two values are equal, and you use = (single equals) when naming arguments that you are passing to functions). Try changing it to use filter(year = 2007) and see what happens.
Grouping rows using group_by()
We see that the life expectancy in 2007 is much larger than the value we got using all of the rows. It seems life expectancy is increasing which is good news. But now we might be interested in calculating the average for each year. Rather that doing a bunch of different filter() statements, we can instead use the group_by() function. The function allows us to tell the code to treat the rows in logical groups, so rather than summarizing over all the rows, we will get one summary value for each group. Here’s what that will look like:
gapminder_data %>%
group_by(year) %>%
summarize(average=mean(lifeExp))
# A tibble: 12 × 2
year average
<dbl> <dbl>
1 1952 49.1
2 1957 51.5
3 1962 53.6
4 1967 55.7
5 1972 57.6
6 1977 59.6
7 1982 61.5
8 1987 63.2
9 1992 64.2
10 1997 65.0
11 2002 65.7
12 2007 67.0
The group_by() function expects you to pass in the name of a column (or multiple columns separated by comma) in your data.
Exercise: Grouping the data
Try calculating the average life expectancy by continent.
Solution
gapminder_data %>% group_by(continent) %>% summarize(average=mean(lifeExp))# A tibble: 5 × 2 continent average <chr> <dbl> 1 Africa 48.9 2 Americas 64.7 3 Asia 60.1 4 Europe 71.9 5 Oceania 74.3By combining
group_by()andsummarize()we are able to calculate the mean life expectancy by continent.
You can also create more than one new column when you call summarize(). To do so, you must separate your columns with a comma. Building on the code from the last exercise, let’s add a new column that calculates the minimum life expectancy for each continent.
gapminder_data %>%
group_by(continent) %>%
summarize(average=mean(lifeExp), min=min(lifeExp))
# A tibble: 5 × 3
continent average min
<chr> <dbl> <dbl>
1 Africa 48.9 23.6
2 Americas 64.7 37.6
3 Asia 60.1 28.8
4 Europe 71.9 43.6
5 Oceania 74.3 69.1
Make new variables with mutate()
Each time we ran summarize(), we got back fewer rows than passed in. We either got one row back, or one row per group. But sometimes we want to create a new column in our data without changing the number of rows. The function we use to create new columns is called mutate().
We have a column for the population and the GDP per capita. If we wanted to get the total GDP, we could multiply the per capita GDP values by the total population. Here’s what such a mutate() command would look like:
gapminder_data %>%
mutate(gdp = pop * gdpPercap)
# A tibble: 1,704 × 7
country year pop continent lifeExp gdpPercap gdp
<chr> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
1 Afghanistan 1952 8425333 Asia 28.8 779. 6567086330.
2 Afghanistan 1957 9240934 Asia 30.3 821. 7585448670.
3 Afghanistan 1962 10267083 Asia 32.0 853. 8758855797.
4 Afghanistan 1967 11537966 Asia 34.0 836. 9648014150.
5 Afghanistan 1972 13079460 Asia 36.1 740. 9678553274.
6 Afghanistan 1977 14880372 Asia 38.4 786. 11697659231.
7 Afghanistan 1982 12881816 Asia 39.9 978. 12598563401.
8 Afghanistan 1987 13867957 Asia 40.8 852. 11820990309.
9 Afghanistan 1992 16317921 Asia 41.7 649. 10595901589.
10 Afghanistan 1997 22227415 Asia 41.8 635. 14121995875.
# … with 1,694 more rows
This will add a new column called “gdp” to our data. We use the column names as if they were regular values that we want to perform mathematical operations on and provide the name in front of an equals sign like we have done with summarize()
Exercise:
mutate()We can also multiply by constants or other numbers using mutate - remember how in the plotting lesson we made a plot with population in millions? Try making a new column for this dataframe called popInMillions that is the population in million.
Solution:
gapminder_data %>% mutate(gdp = pop * gdpPercap, popInMillions = pop / 1000000)# A tibble: 1,704 × 8 country year pop continent lifeExp gdpPercap gdp popInMillions <chr> <dbl> <dbl> <chr> <dbl> <dbl> <dbl> <dbl> 1 Afghanistan 1952 8425333 Asia 28.8 779. 6.57e 9 8.43 2 Afghanistan 1957 9240934 Asia 30.3 821. 7.59e 9 9.24 3 Afghanistan 1962 10267083 Asia 32.0 853. 8.76e 9 10.3 4 Afghanistan 1967 11537966 Asia 34.0 836. 9.65e 9 11.5 5 Afghanistan 1972 13079460 Asia 36.1 740. 9.68e 9 13.1 6 Afghanistan 1977 14880372 Asia 38.4 786. 1.17e10 14.9 7 Afghanistan 1982 12881816 Asia 39.9 978. 1.26e10 12.9 8 Afghanistan 1987 13867957 Asia 40.8 852. 1.18e10 13.9 9 Afghanistan 1992 16317921 Asia 41.7 649. 1.06e10 16.3 10 Afghanistan 1997 22227415 Asia 41.8 635. 1.41e10 22.2 # … with 1,694 more rows
Subset columns using select()
We use the filter() function to choose a subset of the rows from our data, but when we want to choose a subset of columns from our data we use select(). For example, if we only wanted to see the population (“pop”) and year values, we can do:
gapminder_data %>%
select(pop, year)
# A tibble: 1,704 × 2
pop year
<dbl> <dbl>
1 8425333 1952
2 9240934 1957
3 10267083 1962
4 11537966 1967
5 13079460 1972
6 14880372 1977
7 12881816 1982
8 13867957 1987
9 16317921 1992
10 22227415 1997
# … with 1,694 more rows
We can also use select() to drop/remove particular columns by putting a minus sign (-) in front of the column name. For example, if we want everything but the continent column, we can do:
gapminder_data %>%
select(-continent)
# A tibble: 1,704 × 5
country year pop lifeExp gdpPercap
<chr> <dbl> <dbl> <dbl> <dbl>
1 Afghanistan 1952 8425333 28.8 779.
2 Afghanistan 1957 9240934 30.3 821.
3 Afghanistan 1962 10267083 32.0 853.
4 Afghanistan 1967 11537966 34.0 836.
5 Afghanistan 1972 13079460 36.1 740.
6 Afghanistan 1977 14880372 38.4 786.
7 Afghanistan 1982 12881816 39.9 978.
8 Afghanistan 1987 13867957 40.8 852.
9 Afghanistan 1992 16317921 41.7 649.
10 Afghanistan 1997 22227415 41.8 635.
# … with 1,694 more rows
Exercise: selecting columns
Create a dataframe with only the
country,continent,year, andlifeExpcolumns.Solution:
There are multiple ways to do this exercise. Here are two different possibilities.
gapminder_data %>% select(country, continent, year, lifeExp)# A tibble: 1,704 × 4 country continent year lifeExp <chr> <chr> <dbl> <dbl> 1 Afghanistan Asia 1952 28.8 2 Afghanistan Asia 1957 30.3 3 Afghanistan Asia 1962 32.0 4 Afghanistan Asia 1967 34.0 5 Afghanistan Asia 1972 36.1 6 Afghanistan Asia 1977 38.4 7 Afghanistan Asia 1982 39.9 8 Afghanistan Asia 1987 40.8 9 Afghanistan Asia 1992 41.7 10 Afghanistan Asia 1997 41.8 # … with 1,694 more rowsgapminder_data %>% select(-pop, -gdpPercap)# A tibble: 1,704 × 4 country year continent lifeExp <chr> <dbl> <chr> <dbl> 1 Afghanistan 1952 Asia 28.8 2 Afghanistan 1957 Asia 30.3 3 Afghanistan 1962 Asia 32.0 4 Afghanistan 1967 Asia 34.0 5 Afghanistan 1972 Asia 36.1 6 Afghanistan 1977 Asia 38.4 7 Afghanistan 1982 Asia 39.9 8 Afghanistan 1987 Asia 40.8 9 Afghanistan 1992 Asia 41.7 10 Afghanistan 1997 Asia 41.8 # … with 1,694 more rows
Bonus: Using helper functions with
select()The
select()function has a bunch of helper functions that are handy if you are working with a dataset that has a lot of columns. You can see these helper functions on the?selecthelp page. For example, let’s say we wanted to select the year column and all the columns that start with the letter “c”. You can do that with:gapminder_data %>% select(year, starts_with("c"))# A tibble: 1,704 × 3 year country continent <dbl> <chr> <chr> 1 1952 Afghanistan Asia 2 1957 Afghanistan Asia 3 1962 Afghanistan Asia 4 1967 Afghanistan Asia 5 1972 Afghanistan Asia 6 1977 Afghanistan Asia 7 1982 Afghanistan Asia 8 1987 Afghanistan Asia 9 1992 Afghanistan Asia 10 1997 Afghanistan Asia # … with 1,694 more rowsThis returns just the three columns we are interested in.
Exercise: Using
select()with a helper functionFind a helper function on the help page that will choose all the columns that have “p” as their last letter (ie: “pop”,”lifeExp”,”gdpPerCap”)
Solution
The helper function
ends_with()can help us here.gapminder_data %>% select(ends_with("p"))# A tibble: 1,704 × 3 pop lifeExp gdpPercap <dbl> <dbl> <dbl> 1 8425333 28.8 779. 2 9240934 30.3 821. 3 10267083 32.0 853. 4 11537966 34.0 836. 5 13079460 36.1 740. 6 14880372 38.4 786. 7 12881816 39.9 978. 8 13867957 40.8 852. 9 16317921 41.7 649. 10 22227415 41.8 635. # … with 1,694 more rows
Changing the shape of the data
Data comes in many shapes and sizes, and one way we classify data is either “wide” or “long.” Data that is “long” has one row per observation. The gapminder_data data is in a long format. We have one row for each country for each year and each different measurement for that country is in a different column. We might describe this data as “tidy” because it makes it easy to work with ggplot2 and dplyr functions (this is where the “tidy” in “tidyverse” comes from). As tidy as it may be, sometimes we may want our data in a “wide” format. Typically in “wide” format each row represents a group of observations and each value is placed in a different column rather than a different row. For example maybe we want only one row per country and want to spread the life expectancy values into different columns (one for each year).
The tidyr package contains the functions pivot_wider and pivot_longer that make it easy to switch between the two formats. The tidyr package is included in the tidyverse package so we don’t need to do anything to load it.
gapminder_data %>%
select(country, continent, year, lifeExp) %>%
pivot_wider(names_from = year, values_from = lifeExp )
# A tibble: 142 × 14
country continent `1952` `1957` `1962` `1967` `1972` `1977` `1982` `1987`
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Afghanistan Asia 28.8 30.3 32.0 34.0 36.1 38.4 39.9 40.8
2 Albania Europe 55.2 59.3 64.8 66.2 67.7 68.9 70.4 72
3 Algeria Africa 43.1 45.7 48.3 51.4 54.5 58.0 61.4 65.8
4 Angola Africa 30.0 32.0 34 36.0 37.9 39.5 39.9 39.9
5 Argentina Americas 62.5 64.4 65.1 65.6 67.1 68.5 69.9 70.8
6 Australia Oceania 69.1 70.3 70.9 71.1 71.9 73.5 74.7 76.3
7 Austria Europe 66.8 67.5 69.5 70.1 70.6 72.2 73.2 74.9
8 Bahrain Asia 50.9 53.8 56.9 59.9 63.3 65.6 69.1 70.8
9 Bangladesh Asia 37.5 39.3 41.2 43.5 45.3 46.9 50.0 52.8
10 Belgium Europe 68 69.2 70.2 70.9 71.4 72.8 73.9 75.4
# … with 132 more rows, and 4 more variables: 1992 <dbl>, 1997 <dbl>,
# 2002 <dbl>, 2007 <dbl>
Notice here that we tell pivot_wider() which columns to pull the names we wish our new columns to be named from the year variable, and the values to populate those columns from the lifeExp variable. (Again, neither of which have to be in quotes in the code when there are no special characters or spaces - certainly an incentive not to use special characters or spaces!) We see that the resulting tables have new columns by year, and the values populate it with our remaining variables dictating the rows.
Before we move on to more data cleaning, let’s create the final gapminder dataframe we will be working with for the rest of the lesson!
Exercise: final Americas 2007 gapminder dataset
Read in the
gapminder_data.csvfile, filter out the year 2007 and the continent “Americas.” Then drop theyearandcontinentcolumns from the dataframe. Then save the new dataframe into a variable calledgapminder_data_2007.Solution:
gapminder_data_2007 <- read_csv("data/gapminder_data.csv") %>% filter(year == 2007 & continent == "Americas") %>% select(-year, -continent)Rows: 1704 Columns: 6── Column specification ────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────── Delimiter: "," chr (2): country, continent dbl (4): year, pop, lifeExp, gdpPercapℹ Use `spec()` to retrieve the full column specification for this data. ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
Awesome! This is the dataframe we will be using later on in this lesson.
Reviewing Git and GitHub
Now that we have our gapminder data prepared, let’s use what we learned about git and GitHub in the previous lesson to add, commit, and push our changes.
Open Terminal/Git Bash, if you do not have it open already. First we’ll need to navigate to our un-report directory.
Let’s start by print our current working directory and listing the items in the directory, to see where we are.
pwd
ls
Now, we’ll navigate to the un-report directory.
cd ~/Desktop/un-report
ls
To start, let’s pull to make sure our local repository is up to date.
git status
git pull
Not let’s add and commit our changes.
git status
git add
git status "un_data_analysis.R"
git commit -m "Create data analysis file"
Finally, let’s check our commits and then push the commits to GitHub.
git status
git log --online
git push
git status
Cleaning up data
Researchers are often pulling data from several sources, and the process of making data compatible with one another and prepared for analysis can be a large undertaking. Luckily, there are many functions that allow us to do this in R. We’ve been working with the gapminder dataset, which contains population and GDP data by year. In this section, we practice cleaning and preparing a second dataset containing CO2 emissions data by country and year, sourced from the UN.
It’s always good to go into data cleaning with a clear goal in mind. Here, we’d like to prepare the CO2 UN data to be compatible with our gapminder data so we can directly compare GDP to CO2 emissions. To make this work, we’d like a data frame that contains a column with the country name, and columns for different ways of measuring CO2 emissions. We will also want the data to be collected as close to 2007 as possible (the last year we have data for in gapminder). Let’s start with reading the data in using read_csv()
read_csv("data/co2-un-data.csv")
New names:
* `` -> ...3
* `` -> ...4
* `` -> ...5
* `` -> ...6
* `` -> ...7
Rows: 2133 Columns: 7
── Column specification ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
Delimiter: ","
chr (7): T24, CO2 emission estimates, ...3, ...4, ...5, ...6, ...7
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
# A tibble: 2,133 × 7
T24 `CO2 emission estimates` ...3 ...4 ...5 ...6 ...7
<chr> <chr> <chr> <chr> <chr> <chr> <chr>
1 Region/Country/Area <NA> Year Series Value Foot… Source
2 8 Albania 1975 Emissi… 4338… <NA> Inter…
3 8 Albania 1985 Emissi… 6929… <NA> Inter…
4 8 Albania 1995 Emissi… 1848… <NA> Inter…
5 8 Albania 2005 Emissi… 3825… <NA> Inter…
6 8 Albania 2010 Emissi… 3930… <NA> Inter…
7 8 Albania 2015 Emissi… 3824… <NA> Inter…
8 8 Albania 2016 Emissi… 3674… <NA> Inter…
9 8 Albania 2017 Emissi… 4342… <NA> Inter…
10 8 Albania 1975 Emissi… 1.80… <NA> Inter…
# … with 2,123 more rows
The output gives us a warning about missing column names being filled in with things like ‘X3’, ‘X4’, etc. Looking at the table that is outputted by read_csv() we can see that there appear to be two rows at the top of the file that contain information about the data in the table. The first is a header that tells us the table number and its name. Ideally, we’d skip that. We can do this using the skip= argument in read_csv by giving it a number of lines to skip.
read_csv("data/co2-un-data.csv", skip=1)
New names:
* `` -> ...2
Rows: 2132 Columns: 7
── Column specification ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
Delimiter: ","
chr (4): ...2, Series, Footnotes, Source
dbl (3): Region/Country/Area, Year, Value
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
# A tibble: 2,132 × 7
`Region/Country/Area` ...2 Year Series Value Footnotes Source
<dbl> <chr> <dbl> <chr> <dbl> <chr> <chr>
1 8 Albania 1975 Emissions … 4.34e3 <NA> Internation…
2 8 Albania 1985 Emissions … 6.93e3 <NA> Internation…
3 8 Albania 1995 Emissions … 1.85e3 <NA> Internation…
4 8 Albania 2005 Emissions … 3.83e3 <NA> Internation…
5 8 Albania 2010 Emissions … 3.93e3 <NA> Internation…
6 8 Albania 2015 Emissions … 3.82e3 <NA> Internation…
7 8 Albania 2016 Emissions … 3.67e3 <NA> Internation…
8 8 Albania 2017 Emissions … 4.34e3 <NA> Internation…
9 8 Albania 1975 Emissions … 1.80e0 <NA> Internation…
10 8 Albania 1985 Emissions … 2.34e0 <NA> Internation…
# … with 2,122 more rows
Now we get a similar Warning message as before, but the outputted table looks better.
Warnings and Errors:
It’s important to differentiate between Warnings and Errors in R.
- A warning tells us, “you might want to know about this issue, but R still did what you asked”.
- An error tells us, “there’s something wrong with your code or your data and R didn’t do what you asked”.
You need to fix any errors that arise. Warnings, are probably best to resolve or at least understand why they are coming up.
We can resolve this warning by telling read_csv() what the column names should be with the col_names() argument where we give it the column names we want within the c() function separated by commas. If we do this, then we need to set skip to 2 to also skip the column headings.
read_csv("data/co2-un-data.csv", skip=2,
col_names=c("region", "country", "year", "series", "value", "footnotes", "source"))
Rows: 2132 Columns: 7
── Column specification ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
Delimiter: ","
chr (4): country, series, footnotes, source
dbl (3): region, year, value
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
# A tibble: 2,132 × 7
region country year series value footnotes source
<dbl> <chr> <dbl> <chr> <dbl> <chr> <chr>
1 8 Albania 1975 Emissions (thous… 4.34e3 <NA> International Energy…
2 8 Albania 1985 Emissions (thous… 6.93e3 <NA> International Energy…
3 8 Albania 1995 Emissions (thous… 1.85e3 <NA> International Energy…
4 8 Albania 2005 Emissions (thous… 3.83e3 <NA> International Energy…
5 8 Albania 2010 Emissions (thous… 3.93e3 <NA> International Energy…
6 8 Albania 2015 Emissions (thous… 3.82e3 <NA> International Energy…
7 8 Albania 2016 Emissions (thous… 3.67e3 <NA> International Energy…
8 8 Albania 2017 Emissions (thous… 4.34e3 <NA> International Energy…
9 8 Albania 1975 Emissions per ca… 1.80e0 <NA> International Energy…
10 8 Albania 1985 Emissions per ca… 2.34e0 <NA> International Energy…
# … with 2,122 more rows
Bonus: Another way to deal with this error
There are often multiple ways to clean data. Here we read in the table, get the warning and then fix the column names using the rename function.
read_csv("data/co2-un-data.csv", skip=1) %>% rename(country=`...2`)New names: * `` -> ...2Rows: 2132 Columns: 7── Column specification ────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────── Delimiter: "," chr (4): ...2, Series, Footnotes, Source dbl (3): Region/Country/Area, Year, Valueℹ Use `spec()` to retrieve the full column specification for this data. ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.# A tibble: 2,132 × 7 `Region/Country/Area` country Year Series Value Footnotes Source <dbl> <chr> <dbl> <chr> <dbl> <chr> <chr> 1 8 Albania 1975 Emissions … 4.34e3 <NA> Internation… 2 8 Albania 1985 Emissions … 6.93e3 <NA> Internation… 3 8 Albania 1995 Emissions … 1.85e3 <NA> Internation… 4 8 Albania 2005 Emissions … 3.83e3 <NA> Internation… 5 8 Albania 2010 Emissions … 3.93e3 <NA> Internation… 6 8 Albania 2015 Emissions … 3.82e3 <NA> Internation… 7 8 Albania 2016 Emissions … 3.67e3 <NA> Internation… 8 8 Albania 2017 Emissions … 4.34e3 <NA> Internation… 9 8 Albania 1975 Emissions … 1.80e0 <NA> Internation… 10 8 Albania 1985 Emissions … 2.34e0 <NA> Internation… # … with 2,122 more rowsMany data analysts prefer to have their column headings and variable names be in all lower case. We can use a variation of
rename(), which isrename_all()that allows us to set all of the column headings to lower case by giving it the name of the tolower function, which makes everything lowercase.read_csv("data/co2-un-data.csv", skip=1) %>% rename_all(tolower)New names: * `` -> ...2Rows: 2132 Columns: 7── Column specification ────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────── Delimiter: "," chr (4): ...2, Series, Footnotes, Source dbl (3): Region/Country/Area, Year, Valueℹ Use `spec()` to retrieve the full column specification for this data. ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.# A tibble: 2,132 × 7 `region/country/area` ...2 year series value footnotes source <dbl> <chr> <dbl> <chr> <dbl> <chr> <chr> 1 8 Albania 1975 Emissions … 4.34e3 <NA> Internation… 2 8 Albania 1985 Emissions … 6.93e3 <NA> Internation… 3 8 Albania 1995 Emissions … 1.85e3 <NA> Internation… 4 8 Albania 2005 Emissions … 3.83e3 <NA> Internation… 5 8 Albania 2010 Emissions … 3.93e3 <NA> Internation… 6 8 Albania 2015 Emissions … 3.82e3 <NA> Internation… 7 8 Albania 2016 Emissions … 3.67e3 <NA> Internation… 8 8 Albania 2017 Emissions … 4.34e3 <NA> Internation… 9 8 Albania 1975 Emissions … 1.80e0 <NA> Internation… 10 8 Albania 1985 Emissions … 2.34e0 <NA> Internation… # … with 2,122 more rows
We previously saw how we can subset columns from a data frame using the select function. There are a lot of columns with extraneous information in this dataset, let’s subset out the columns we are interested in.
Exercise: reviewing selecting columns
Select the country, year, series, and value columns from our dataset.
Solution:
read_csv("data/co2-un-data.csv", skip=2, col_names=c("region", "country", "year", "series", "value", "footnotes", "source")) %>% select(country, year, series, value)Rows: 2132 Columns: 7── Column specification ────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────── Delimiter: "," chr (4): country, series, footnotes, source dbl (3): region, year, valueℹ Use `spec()` to retrieve the full column specification for this data. ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.# A tibble: 2,132 × 4 country year series value <chr> <dbl> <chr> <dbl> 1 Albania 1975 Emissions (thousand metric tons of carbon dioxide) 4338. 2 Albania 1985 Emissions (thousand metric tons of carbon dioxide) 6930. 3 Albania 1995 Emissions (thousand metric tons of carbon dioxide) 1849. 4 Albania 2005 Emissions (thousand metric tons of carbon dioxide) 3825. 5 Albania 2010 Emissions (thousand metric tons of carbon dioxide) 3930. 6 Albania 2015 Emissions (thousand metric tons of carbon dioxide) 3825. 7 Albania 2016 Emissions (thousand metric tons of carbon dioxide) 3674. 8 Albania 2017 Emissions (thousand metric tons of carbon dioxide) 4342. 9 Albania 1975 Emissions per capita (metric tons of carbon dioxide) 1.80 10 Albania 1985 Emissions per capita (metric tons of carbon dioxide) 2.34 # … with 2,122 more rows
The series column has two methods of quantifying CO2 emissions - “Emissions (thousand metric tons of carbon dioxide)” and “Emissions per capita (metric tons of carbon dioxide)”. Those are long titles that we’d like to shorten to make them easier to work with. We can shorten them to “total_emissions” and “per_capita_emissions” using the recode function. We need to do this within the mutate function where we will mutate the series column. The syntax in the recode function is to tell recode which column we want to recode and then what the old value (e.g. “Emissions (thousand metric tons of carbon dioxide)”) should equal after recoding (e.g. “total”).
read_csv("data/co2-un-data.csv", skip=2,
col_names=c("region", "country", "year", "series", "value", "footnotes", "source")) %>%
select(country, year, series, value) %>%
mutate(series = recode(series, "Emissions (thousand metric tons of carbon dioxide)" = "total_emissions",
"Emissions per capita (metric tons of carbon dioxide)" = "per_capita_emissions"))
Rows: 2132 Columns: 7
── Column specification ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
Delimiter: ","
chr (4): country, series, footnotes, source
dbl (3): region, year, value
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
# A tibble: 2,132 × 4
country year series value
<chr> <dbl> <chr> <dbl>
1 Albania 1975 total_emissions 4338.
2 Albania 1985 total_emissions 6930.
3 Albania 1995 total_emissions 1849.
4 Albania 2005 total_emissions 3825.
5 Albania 2010 total_emissions 3930.
6 Albania 2015 total_emissions 3825.
7 Albania 2016 total_emissions 3674.
8 Albania 2017 total_emissions 4342.
9 Albania 1975 per_capita_emissions 1.80
10 Albania 1985 per_capita_emissions 2.34
# … with 2,122 more rows
Recall that we’d like to have separate columns for the two ways that we CO2 emissions data. To achieve this, we’ll use the pivot_wider function that we saw previously. The columns we want to spread out are series (i.e. names_from) and value (i.e. values_from).
read_csv("data/co2-un-data.csv", skip=2,
col_names=c("region", "country", "year", "series", "value", "footnotes", "source")) %>%
select(country, year, series, value) %>%
mutate(series = recode(series, "Emissions (thousand metric tons of carbon dioxide)" = "total_emission",
"Emissions per capita (metric tons of carbon dioxide)" = "per_capita_emission")) %>%
pivot_wider(names_from=series, values_from=value)
Rows: 2132 Columns: 7
── Column specification ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
Delimiter: ","
chr (4): country, series, footnotes, source
dbl (3): region, year, value
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
# A tibble: 1,066 × 4
country year total_emission per_capita_emission
<chr> <dbl> <dbl> <dbl>
1 Albania 1975 4338. 1.80
2 Albania 1985 6930. 2.34
3 Albania 1995 1849. 0.58
4 Albania 2005 3825. 1.27
5 Albania 2010 3930. 1.35
6 Albania 2015 3825. 1.33
7 Albania 2016 3674. 1.28
8 Albania 2017 4342. 1.51
9 Algeria 1975 13553. 0.811
10 Algeria 1985 42073. 1.86
# … with 1,056 more rows
Excellent! The last step before we can join this data frame is to get the most data that is for the year closest to 2007 so we can make a more direct comparison to the most recent data we have from gapminder. For the sake of time, we’ll just tell you that we want data from 2005.
Bonus: How did we determine that 2005 is the closest year to 2007?
We want to make sure we pick a year that is close to 2005, but also a year that has a decent amount of data to work with. One useful tool is the
count()function, which will tell us how many times a value is repeated in a column of a data frame. Let’s use this function on the year column to see which years we have data for and to tell us whether we have a good number of countries represented in that year.read_csv("data/co2-un-data.csv", skip=2, col_names=c("region", "country", "year", "series", "value", "footnotes", "source")) %>% select(country, year, series, value) %>% mutate(series = recode(series, "Emissions (thousand metric tons of carbon dioxide)" = "total", "Emissions per capita (metric tons of carbon dioxide)" = "per_capita")) %>% pivot_wider(names_from=series, values_from=value) %>% count(year)Rows: 2132 Columns: 7── Column specification ────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────── Delimiter: "," chr (4): country, series, footnotes, source dbl (3): region, year, valueℹ Use `spec()` to retrieve the full column specification for this data. ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.# A tibble: 8 × 2 year n <dbl> <int> 1 1975 111 2 1985 113 3 1995 136 4 2005 140 5 2010 140 6 2015 142 7 2016 142 8 2017 142It looks like we have data for 140 countries in 2005 and 2010. We chose 2005 because it is closer to 2007.
Exercise:
Filter out data from 2005 and then drop the year column. (Since we will have only data from one year, it is now irrelevant.)
Solution:
read_csv("data/co2-un-data.csv", skip=2, col_names=c("region", "country", "year", "series", "value", "footnotes", "source")) %>% select(country, year, series, value) %>% mutate(series = recode(series, "Emissions (thousand metric tons of carbon dioxide)" = "total", "Emissions per capita (metric tons of carbon dioxide)" = "per_capita")) %>% pivot_wider(names_from=series, values_from=value) %>% filter(year==2005) %>% select(-year)Rows: 2132 Columns: 7── Column specification ────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────── Delimiter: "," chr (4): country, series, footnotes, source dbl (3): region, year, valueℹ Use `spec()` to retrieve the full column specification for this data. ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.# A tibble: 140 × 3 country total per_capita <chr> <dbl> <dbl> 1 Albania 3825. 1.27 2 Algeria 77474. 2.33 3 Angola 6147. 0.314 4 Argentina 149476. 3.82 5 Armenia 4130. 1.38 6 Australia 365515. 17.9 7 Austria 74764. 9.09 8 Azerbaijan 29018. 3.46 9 Bahrain 20565. 23.1 10 Bangladesh 31960. 0.223 # … with 130 more rows
Finally, let’s go ahead and assign the output of this code chunk to a variable name:
co2_emissions <- read_csv("data/co2-un-data.csv", skip=2,
col_names=c("region", "country", "year", "series", "value", "footnotes", "source")) %>%
select(country, year, series, value) %>%
mutate(series = recode(series, "Emissions (thousand metric tons of carbon dioxide)" = "total_emission",
"Emissions per capita (metric tons of carbon dioxide)" = "per_capita_emission")) %>%
pivot_wider(names_from=series, values_from=value) %>%
filter(year==2005) %>%
select(-year)
Rows: 2132 Columns: 7
── Column specification ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
Delimiter: ","
chr (4): country, series, footnotes, source
dbl (3): region, year, value
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
Looking at your data
You can get a look at your data-cleaning hard work by navigating to the Environment tab in RStudio and clicking the table icon next to the variable name. Notice when we do this, RStudio automatically runs the
View()command. We’ve made a lot of progress!
Joining data frames
Now we’re ready to join our CO2 emissions data to the gapminder data. Previously we saw that we could read in and filter the gapminder data like this to get the data from the Americas for 2007 so we can create a new dataframe with our filtered data:
gapminder_data_2007 <- read_csv("data/gapminder_data.csv") %>%
filter(year == 2007 & continent == "Americas") %>%
select(-year, -continent)
Rows: 1704 Columns: 6
── Column specification ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
Delimiter: ","
chr (2): country, continent
dbl (4): year, pop, lifeExp, gdpPercap
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
Look at the data in co2_emissions and gapminder_data_2007. If you had to merge these two data frames together, which column would you use to merge them together? If you said “country” - good job!
We’ll call country our “key”. Now, when we join them together, can you think of any problems we might run into when we merge things? We might not have CO2 emissions data for all of the countries in the gapminder dataset and vice versa. Also, a country might be represented in both data frames but not by the same name in both places.
The dplyr package has a number of tools for joining data frames together depending on what we want to do with the rows of the data of countries that are not represented in both data frames. Here we’ll be using inner_join() and anti_join().
In an “inner join”, the new data frame only has those rows where the same key is found in both data frames. This is a very commonly used join.

Bonus: Other dplyr join functions
Outer joins and can be performed using
left_join(),right_join(), andfull_join(). In a “left join”, if the key is present in the left hand data frame, it will appear in the output, even if it is not found in the the right hand data frame. For a right join, the opposite is true. For a full join, all possible keys are included in the output data frame.
Let’s give the inner_join() function a try.
inner_join(gapminder_data, co2_emissions)
Joining, by = "country"
# A tibble: 1,188 × 8
country year pop continent lifeExp gdpPercap total_emission
<chr> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
1 Albania 1952 1282697 Europe 55.2 1601. 3825.
2 Albania 1957 1476505 Europe 59.3 1942. 3825.
3 Albania 1962 1728137 Europe 64.8 2313. 3825.
4 Albania 1967 1984060 Europe 66.2 2760. 3825.
5 Albania 1972 2263554 Europe 67.7 3313. 3825.
6 Albania 1977 2509048 Europe 68.9 3533. 3825.
7 Albania 1982 2780097 Europe 70.4 3631. 3825.
8 Albania 1987 3075321 Europe 72 3739. 3825.
9 Albania 1992 3326498 Europe 71.6 2497. 3825.
10 Albania 1997 3428038 Europe 73.0 3193. 3825.
# … with 1,178 more rows, and 1 more variable: per_capita_emission <dbl>
Do you see that we now have data from both data frames joined together in the same data frame? One thing to note about the output is that inner_join() tells us that that it joined by “country”. We can make this explicit using the “by” argument in the join functions
inner_join(gapminder_data, co2_emissions, by="country")
# A tibble: 1,188 × 8
country year pop continent lifeExp gdpPercap total_emission
<chr> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
1 Albania 1952 1282697 Europe 55.2 1601. 3825.
2 Albania 1957 1476505 Europe 59.3 1942. 3825.
3 Albania 1962 1728137 Europe 64.8 2313. 3825.
4 Albania 1967 1984060 Europe 66.2 2760. 3825.
5 Albania 1972 2263554 Europe 67.7 3313. 3825.
6 Albania 1977 2509048 Europe 68.9 3533. 3825.
7 Albania 1982 2780097 Europe 70.4 3631. 3825.
8 Albania 1987 3075321 Europe 72 3739. 3825.
9 Albania 1992 3326498 Europe 71.6 2497. 3825.
10 Albania 1997 3428038 Europe 73.0 3193. 3825.
# … with 1,178 more rows, and 1 more variable: per_capita_emission <dbl>
One thing to notice is that gapminder data had 25 rows, but the output of our join only had 21. Let’s investigate. It appears that there must have been countries in the gapminder data that did not appear in our co2_emissions data frame.
Let’s use anti_join() for this - this will show us the data for the keys on the left that are missing from the data frame on the right.
anti_join(gapminder_data, co2_emissions, by="country")
# A tibble: 516 × 6
country year pop continent lifeExp gdpPercap
<chr> <dbl> <dbl> <chr> <dbl> <dbl>
1 Afghanistan 1952 8425333 Asia 28.8 779.
2 Afghanistan 1957 9240934 Asia 30.3 821.
3 Afghanistan 1962 10267083 Asia 32.0 853.
4 Afghanistan 1967 11537966 Asia 34.0 836.
5 Afghanistan 1972 13079460 Asia 36.1 740.
6 Afghanistan 1977 14880372 Asia 38.4 786.
7 Afghanistan 1982 12881816 Asia 39.9 978.
8 Afghanistan 1987 13867957 Asia 40.8 852.
9 Afghanistan 1992 16317921 Asia 41.7 649.
10 Afghanistan 1997 22227415 Asia 41.8 635.
# … with 506 more rows
We can see that the co2_emissions data were missing for Bolivia, Puerto Rico, United States, and Venezuela.
If we look at the co2_emissions data with View(), we will see that Bolivia, United States, and Venezuela are called different things in the co2_emissions data frame. They’re called “Bolivia (Plurin. State of)”, “United States of America”, and “Venezuela (Boliv. Rep. of)”. Puerto Rico isn’t a country; it’s part of the United States. Using mutate() and recode(), we can re-import the co2_emissions data so that the country names for Bolivia, United States, and Venezuela, match those in the gapminder data.
co2_emissions <- read_csv("data/co2-un-data.csv", skip=2,
col_names=c("region", "country", "year",
"series", "value", "footnotes", "source")) %>%
select(country, year, series, value) %>%
mutate(series = recode(series, "Emissions (thousand metric tons of carbon dioxide)" = "total",
"Emissions per capita (metric tons of carbon dioxide)" = "per_capita")) %>%
pivot_wider(names_from=series, values_from=value) %>%
filter(year==2005) %>%
select(-year) %>%
mutate(country=recode(country,
"Bolivia (Plurin. State of)" = "Bolivia",
"United States of America" = "United States",
"Venezuela (Boliv. Rep. of)" = "Venezuela")
)
Rows: 2132 Columns: 7
── Column specification ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
Delimiter: ","
chr (4): country, series, footnotes, source
dbl (3): region, year, value
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
anti_join(gapminder_data, co2_emissions, by="country")
# A tibble: 480 × 6
country year pop continent lifeExp gdpPercap
<chr> <dbl> <dbl> <chr> <dbl> <dbl>
1 Afghanistan 1952 8425333 Asia 28.8 779.
2 Afghanistan 1957 9240934 Asia 30.3 821.
3 Afghanistan 1962 10267083 Asia 32.0 853.
4 Afghanistan 1967 11537966 Asia 34.0 836.
5 Afghanistan 1972 13079460 Asia 36.1 740.
6 Afghanistan 1977 14880372 Asia 38.4 786.
7 Afghanistan 1982 12881816 Asia 39.9 978.
8 Afghanistan 1987 13867957 Asia 40.8 852.
9 Afghanistan 1992 16317921 Asia 41.7 649.
10 Afghanistan 1997 22227415 Asia 41.8 635.
# … with 470 more rows
Now we see that our recode enabled the join for all countries in the gapminder, and we are left with Puerto Rico. In the next exercise, Let’s recode Puerto Rico as United States in the gapminder data and then use group_by() and summarize() to aggregate the data; we’ll use the population data to weight the life expectancy and GDP values.
In the gapminder data, let’s recode Puerto Rico as United States.
gapminder_data <- read_csv("data/gapminder_data.csv") %>%
filter(year == 2007 & continent == "Americas") %>%
select(-year, -continent) %>%
mutate(country = recode(country, "Puerto Rico" = "United States"))
Rows: 1704 Columns: 6
── Column specification ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
Delimiter: ","
chr (2): country, continent
dbl (4): year, pop, lifeExp, gdpPercap
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
Now we have to group Puerto Rico and the US together, aggregating and calculating the data for all of the other columns. This is a little tricky - we will need a weighted average of lifeExp and gdpPercap.
gapminder_data <- read_csv("data/gapminder_data.csv") %>%
filter(year == 2007 & continent == "Americas") %>%
select(-year, -continent) %>%
mutate(country = recode(country, "Puerto Rico" = "United States")) %>%
group_by(country) %>%
summarize(lifeExp = sum(lifeExp * pop)/sum(pop),
gdpPercap = sum(gdpPercap * pop)/sum(pop),
pop = sum(pop)
)
Rows: 1704 Columns: 6
── Column specification ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
Delimiter: ","
chr (2): country, continent
dbl (4): year, pop, lifeExp, gdpPercap
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
Let’s check to see if it worked!
anti_join(gapminder_data, co2_emissions, by="country")
# A tibble: 0 × 4
# … with 4 variables: country <chr>, lifeExp <dbl>, gdpPercap <dbl>, pop <dbl>
Now our anti_join() returns an empty data frame, which tells us that we have reconciled all of the keys from the gapminder data with the data in the co2_emissions data frame.
Finally, let’s use the inner_join() to create a new data frame:
gapminder_co2 <- inner_join(gapminder_data, co2_emissions, by="country")
One last thing! What if we’re interested in distinguishing between countries in North America and South America? We want to create two groups - Canada, the United States, and Mexico in one and the other countries in another.
We can create a grouping variable using mutate() combined with an if_else() function - a very useful pairing.
gapminder_co2 %>%
mutate(region = if_else(country == "Canada" | country == "United States" | country == "Mexico", "north", "south"))
# A tibble: 24 × 7
country lifeExp gdpPercap pop total per_capita region
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <chr>
1 Argentina 75.3 12779. 40301927 149476. 3.82 south
2 Bolivia 65.6 3822. 9119152 8976. 0.984 south
3 Brazil 72.4 9066. 190010647 311624. 1.67 south
4 Canada 80.7 36319. 33390141 540431. 16.8 north
5 Chile 78.6 13172. 16284741 54435. 3.34 south
6 Colombia 72.9 7007. 44227550 53585. 1.24 south
7 Costa Rica 78.8 9645. 4133884 5463. 1.29 south
8 Cuba 78.3 8948. 11416987 25051. 2.22 south
9 Dominican Republic 72.2 6025. 9319622 17522. 1.90 south
10 Ecuador 75.0 6873. 13755680 23927. 1.74 south
# … with 14 more rows
Let’s look at the output - see how the Canada, US, and Mexico rows are all labeled as “north” and everything else is labeled as “south”
We have reached our data cleaning goals! One of the best aspects of doing all of these steps coded in R is that our efforts are reproducible, and the raw data is maintained. With good documentation of data cleaning and analysis steps, we could easily share our work with another researcher who would be able to repeat what we’ve done. However, it’s also nice to have a saved csv copy of our clean data. That way we can access it later without needing to redo our data cleaning, and we can also share the cleaned data with collaborators. To save our dataframe, we’ll use write_csv().
write_csv(gapminder_co2, "data/gapminder_co2.csv")
Great - Now we can move on to the analysis!
Analyzing combined data
For our analysis, we have two questions we’d like to answer. First, is there a relationship between the GDP of a country and the amount of CO2 emitted (per capita)? Second, Canada, the United States, and Mexico account for nearly half of the population of the Americas. What percent of the total CO2 production do they account for?
To answer the first question, we’ll plot the CO2 emitted (on a per capita basis) against the GDP (on a per capita basis) using a scatter plot:
ggplot(gapminder_co2, aes(x=gdpPercap, y=per_capita)) +
geom_point() +
labs(x="GDP (per capita)",
y="CO2 emitted (per capita)",
title="There is a strong association between a nation's GDP \nand the amount of CO2 it produces"
)
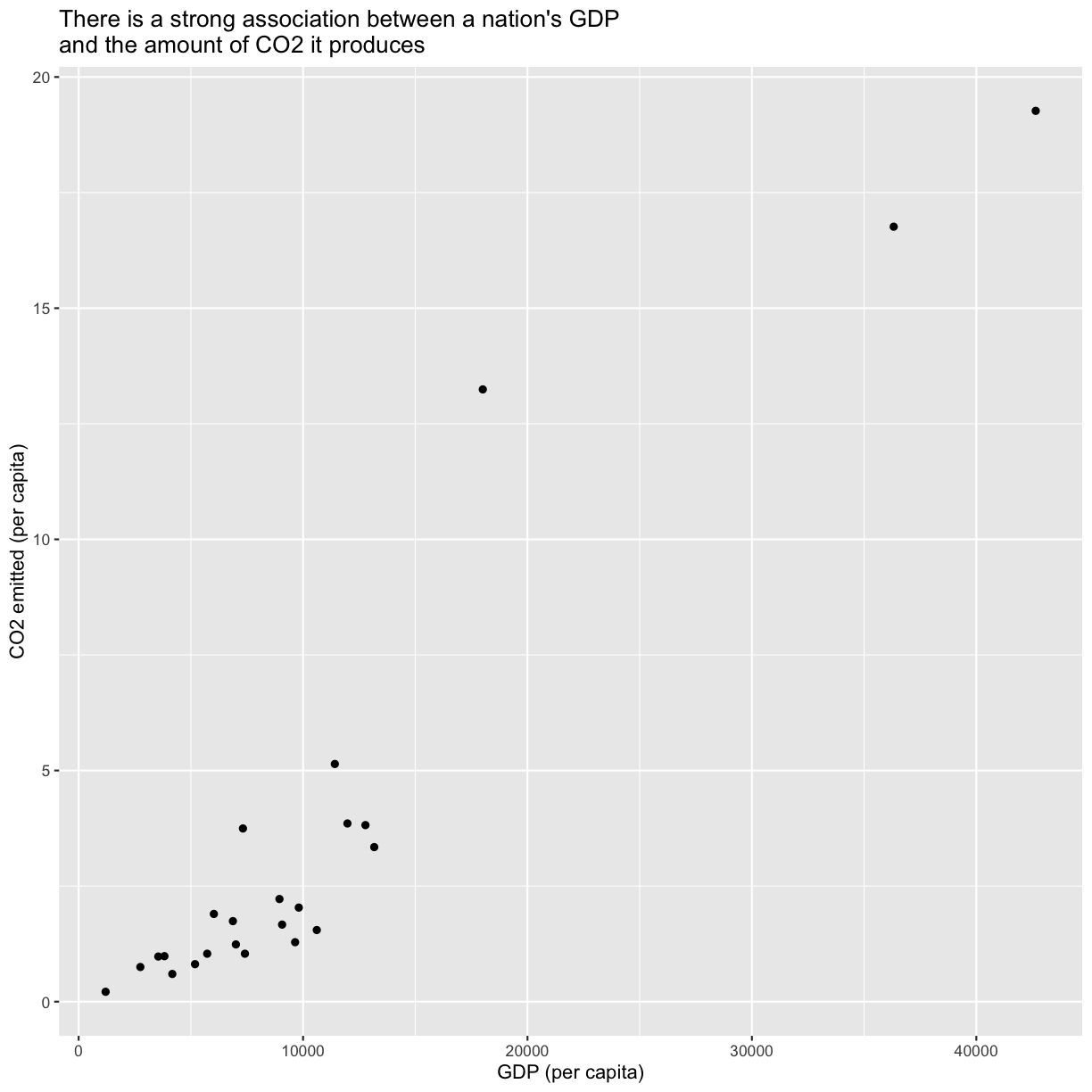
Tip: Notice we used the \n in our title to get a new line to prevent it from getting cut off.
To help clarify the association, we can add a fit line through the data using geom_smooth()
ggplot(gapminder_co2, aes(x=gdpPercap, y=per_capita)) +
geom_point() +
labs(x="GDP (per capita)",
y="CO2 emitted (per capita)",
title="There is a strong association between a nation's GDP \nand the amount of CO2 it produces"
) +
geom_smooth()
`geom_smooth()` using method = 'loess' and formula 'y ~ x'
We can force the line to be straight using method="lm" as an argument to geom_smooth
ggplot(gapminder_co2, aes(x=gdpPercap, y=per_capita)) +
geom_point() +
labs(x="GDP (per capita)",
y="CO2 emitted (per capita)",
title="There is a strong association between a nation's GDP \nand the amount of CO2 it produces"
) +
geom_smooth(method="lm")
`geom_smooth()` using formula 'y ~ x'
To answer our first question, as the title of our plot indicates there is indeed a strong association between a nation’s GDP and the amount of CO2 it produces.
For the second question, we want to create two groups - Canada, the United States, and Mexico in one and the other countries in another.
We can create a grouping variable using mutate() combined with an if_else() function - a very useful pairing.
gapminder_co2 %>%
mutate(region = if_else(country == "Canada" | country == "United States" | country == "Mexico", "north", "south"))
# A tibble: 24 × 7
country lifeExp gdpPercap pop total per_capita region
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <chr>
1 Argentina 75.3 12779. 40301927 149476. 3.82 south
2 Bolivia 65.6 3822. 9119152 8976. 0.984 south
3 Brazil 72.4 9066. 190010647 311624. 1.67 south
4 Canada 80.7 36319. 33390141 540431. 16.8 north
5 Chile 78.6 13172. 16284741 54435. 3.34 south
6 Colombia 72.9 7007. 44227550 53585. 1.24 south
7 Costa Rica 78.8 9645. 4133884 5463. 1.29 south
8 Cuba 78.3 8948. 11416987 25051. 2.22 south
9 Dominican Republic 72.2 6025. 9319622 17522. 1.90 south
10 Ecuador 75.0 6873. 13755680 23927. 1.74 south
# … with 14 more rows
Now we can use this column to repeat our group_by() and summarize() steps
gapminder_co2 %>%
mutate(region = if_else(country == "Canada" |
country == "United States" |
country == "Mexico", "north", "south")) %>%
group_by(region) %>%
summarize(sumtotal = sum(total),
sumpop = sum(pop))
# A tibble: 2 × 3
region sumtotal sumpop
<chr> <dbl> <dbl>
1 north 6656037. 447173470
2 south 889332. 451697714
The if_else() statement reads like, “if country equals “Canada” OR | “United states” OR “Mexico”, The new variable region should be “north”, else “south””. It’s worth exploring logical operators for “or” |, “and” &&, and “not” !, which opens up a great deal of possibilities for writing code to do what you want.
We see that although Canada, the United States, and Mexico account for close to half the population of the Americas, they account for 88% of the CO2 emitted. We just did this math quickly by plugging the numbers from our table into the console to get the percentages. Can we make that a little more reproducible by calculating percentages for population (pop) and total emissions (total) into our data before summarizing?
Finishing with Git and GitHub
Awesome work! Let’s make sure it doesn’t go to waste. Time to add, commit, and push our changes to GitHub again - do you remember how?
Exercise: changing directories
Print your current working directory and list the items in the directory to check where you are. If you are not in the un-report directory, navigate there.
Solution:
pwd ls cd ~/Desktop/un-report ls
Exercise: reviewing git and GitHub
Pull to make sure our local repository is up to date. Then add, commit, and push your commits to GitHub. Don’t forget to check your git status periodically to make sure everything is going as expected!
Solution:
git status git pull git status git add "data-analysis.R" git status git commit -m "Create data analysis file" git status git log --online git push git status
Putting it all together
We’ve gone through many steps of cleaning, joining, and analyzing our data.
As a challenge, create an RScript file that contains the code to clean up the
gapminder and CO2 emissions data, joins the two data frames, and builds the
plot and summary table comparing Canada, the United States, and Mexico to the
rest of the Americas. You can even practice some of the ggplot skills you
learned earlier to move that plot beyond the ggplot2 defaults.
# Load and prepare gapminder data
gapminder_co2 %>%
mutate(region = if_else(country == "Canada" |
country == "United States" |
country == "Mexico", "north", "south")) %>%
group_by(region) %>%
summarize(sumtotal = sum(total),
sumpop = sum(pop))
# A tibble: 2 × 3
region sumtotal sumpop
<chr> <dbl> <dbl>
1 north 6656037. 447173470
2 south 889332. 451697714
# Load and prepare co2 emissions data
co2_emissions <- read_csv("data/co2-un-data.csv", skip=2,
col_names=c("region", "country", "year", "series", "value", "footnotes", "source")) %>%
select(country, year, series, value) %>%
mutate(series = recode(series, "Emissions (thousand metric tons of carbon dioxide)" = "total",
"Emissions per capita (metric tons of carbon dioxide)" = "per_capita")) %>%
pivot_wider(names_from=series, values_from=value) %>%
filter(year==2005) %>%
select(-year) %>%
mutate(country=recode(country,
"Bolivia (Plurin. State of)" = "Bolivia",
"United States of America" = "United States",
"Venezuela (Boliv. Rep. of)" = "Venezuela")
)
Rows: 2132 Columns: 7
── Column specification ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
Delimiter: ","
chr (4): country, series, footnotes, source
dbl (3): region, year, value
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
# Join the two data frames
gapminder_co2 <- inner_join(gapminder_data, co2_emissions, by="country")
# Plot
ggplot(gapminder_co2, aes(x=gdpPercap, y=per_capita)) +
geom_point() +
labs(x="GDP (per capita)",
y="CO2 emitted (per capita)",
title="There is a strong association between a nation's GDP \nand the amount of CO2 it produces"
) +
geom_smooth(method="lm")
`geom_smooth()` using formula 'y ~ x'
# Table
gapminder_co2 %>%
mutate(region = if_else(country == "Canada" | country == "United States" | country == "Mexico", "north", "south")) %>%
mutate(totalPercent = total/sum(total)*100,
popPercent = pop/sum(pop)*100) %>%
group_by(region) %>%
summarize(sumTotalPercent = sum(totalPercent),
sumPopPercent = sum(popPercent))
# A tibble: 2 × 3
region sumTotalPercent sumPopPercent
<chr> <dbl> <dbl>
1 north 88.2 49.7
2 south 11.8 50.3
Key Points
Package loading is an important first step in preparing an R environment.
Data analsyis in R facilitates reproducible research.
There are many useful functions in the
tidyversepackages that can aid in data analysis.Assessing data source and structure is an important first step in analysis.
Preparing data for analysis can take significant effort and planning.
Writing Reports with R Markdown
Overview
Teaching: 105 min
Exercises: 30 minQuestions
How can I make reproducible reports using R Markdown?
How do I format text using Markdown?
Objectives
To create a report in R Markdown that combines text, code, and figures.
To use Markdown to format our report.
To understand how to use R code chunks to include or hide code, figures, and messages.
To be aware of the various report formats that can be rendered using R Markdown.
To practice using the Unix Shell, Github, and R through paired programming exercises.
Contents
- What is R Markdown and why use it?
- Creating a reports directory
- Creating an R Markdown file
- Basic components of R Markdown
- Starting the report
- Formatting
- Integrating it all together: Paired exercise
Recall that our goal is to generate a report to the United Nations on how a country’s life expectancy is related to GDP.
Discusion
How do you usually share data analyses with your collaborators? Many people share them through a Word or PDF document, a spreadsheet, slides, a graphic, etc.
What is R Markdown and why use it?
We’re going to learn a way to create a report in R using something called R Markdown. R Markdown combines Markdown (which renders plain text) with R code, which you can use to do data analysis and make figures. R Markdown enables you to “render” report of our analysis and results into an html, PDF or Word file that you can share with others.
Combining Markdown and R code in the same document allows you to integrate the text of the report, the code you used, and your figures, tables, and other output into one document. The text can be your introduction, description of the research design and experimental methods, your analysis and discussion of the results, the conclusion, or whatever else you want to include. R Markdown reports can be used for many different things, including reproducible analysis reports, publication, sharing work with collaborators, writing up homework, or keeping a computational notebook of your work. (If you are familiar with Jupyter notebooks in the Python programming environment, R Markdown is R’s equivalent of a Jupyter notebook.)
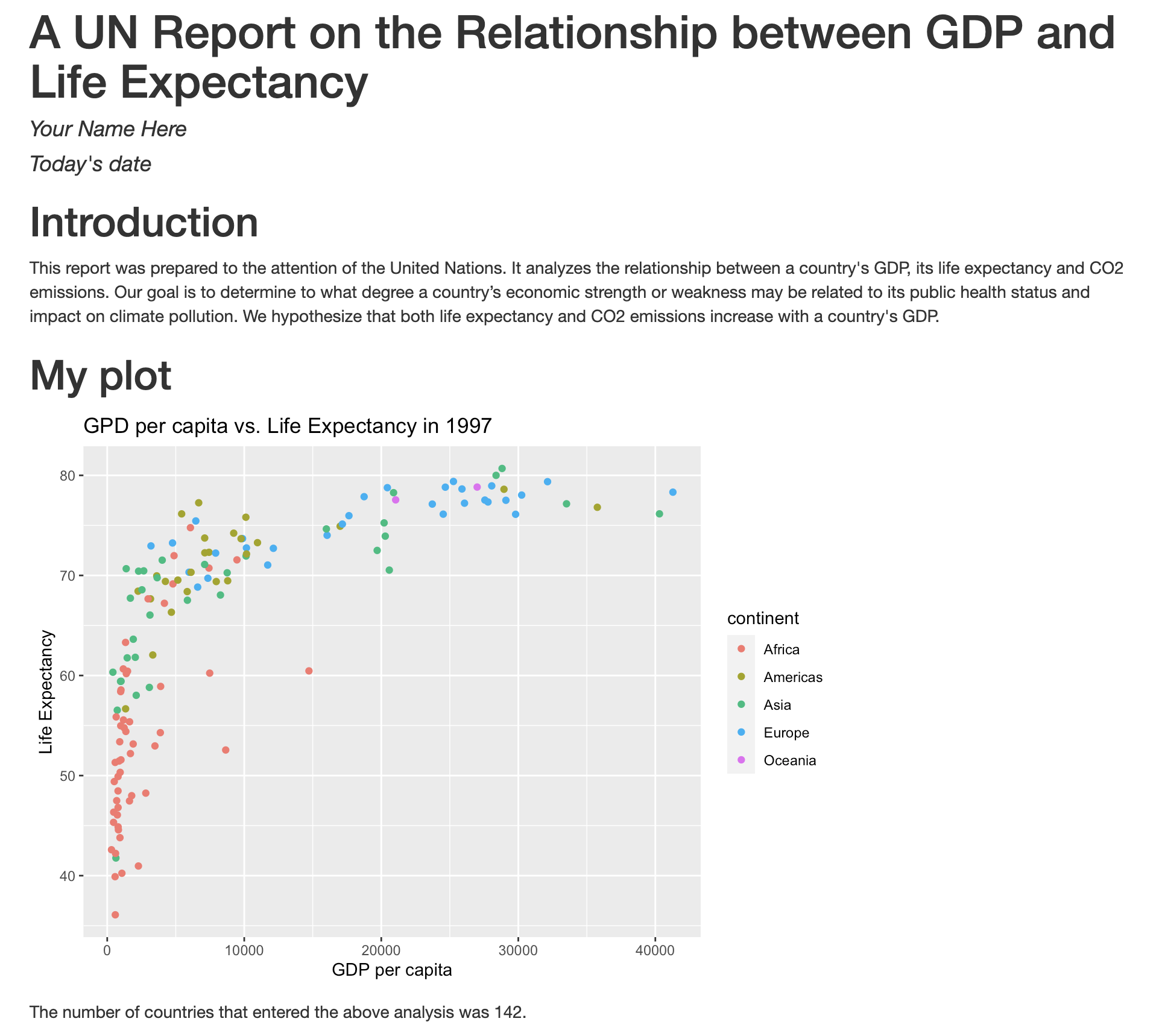
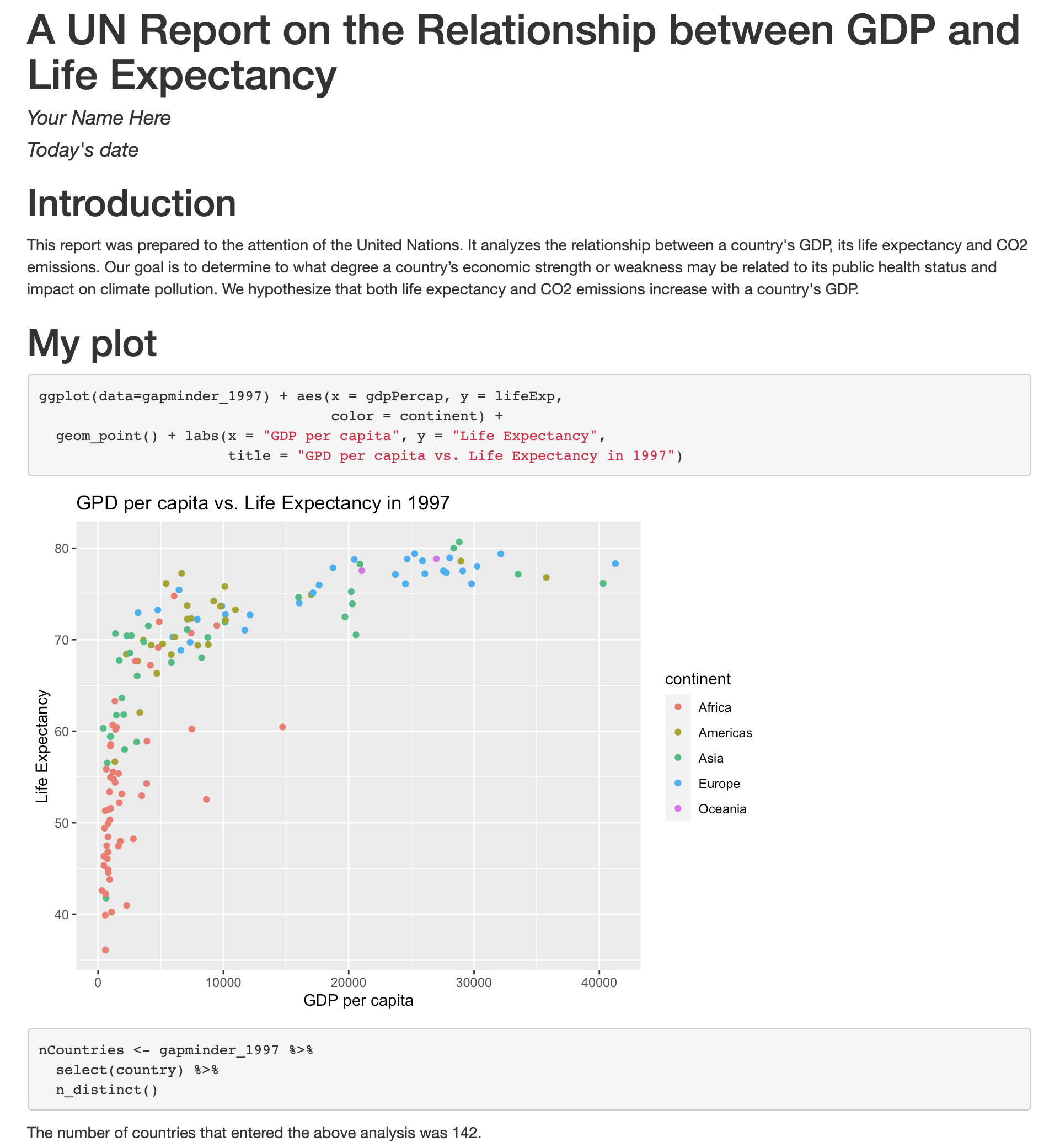
We call reports reproducible when the code is embedded in the document and anyone can take the report and re-run the code to get the same output. If you’re writing a publication or sharing results with collaborators who won’t be interested in seeing that nitty-gritty part of your work, you can hide the code in the final, rendered output file while keeping it in the R Markdown file for your own record. Writing reports with R Markdown is also useful because if you find an error or want to modify the text or a figure later on, you can simply change the code and re-run the R Markdown file to get the updated output!
Creating a reports directory
To get started, let’s use the Unix Shell to create a directory within un-report called reports where we will write our reports to the UN.
First, open the Unix Shell and cd to un-report
pwd
mkdir reports
/home/USERNAME/Desktop/un-report/reports/
Creating an R Markdown file
Now that we have a better understanding of what we can use R Markdown files for, let’s start writing a report!
To create an R Markdown file:
- Open RStudio
- Go to File → New File → R Markdown
- Give your document a title, something like “A UN Report on the Relationship between GDP and Life Expectancy” (Note: this is not the same as the file name - it’s just a title that will appear at the top of your report)
- Keep the default output format as HTML.
- R Markdown files always end in
.Rmd
R Markdown Outputs
The default output for an R Markdown report is HTML, but you can also use R Markdown to output other report formats. For example, you can generate PDF reports using R Markdown, but you must install TeX to do this.
Basic components of R Markdown
Header
The first part is a header at the top of the file between the lines of ---. This contains instructions for R to specify the type of document to be created and options to choose (ex., title, author, date). These are in the form of key-value pairs (key: value; YAML).
Here’s an example:
---
title: 'Writing Reports with R Markdown'
author: "Zena Lapp"
date: "11/18/2020"
output: html_document
---
Code chunks
The next section is a code chunk, or embedded R code, that sets up options for all code chunks. Here is the default when you create a new R Markdown file:
```{r setup, include=FALSE}
knitr::opts_knit$set(root.dir = normalizePath("..")
```
```{r setup, include=FALSE}
knitr::opts_chunk$set(echo = TRUE)
```
All code chunks have this format:
```{r}
# Your code here
```
All of the code is enclosed in 3 backticks (cat('``’)), and the {r}` part indicates that it’s a chunk of R code.
You can also include other information within the curly brackets to indicate different information about that code chunk.
For instance, the first code block is named “setup”, and include=FALSE prevents code and results from showing up in the output file.
Inside the code chunk, you can put any R code that you want to run, and you can have as many code chunks as you want in your file.
As we mentioned above, in the first code chunk you set options for the entire file.
echo = TRUE means that you want your code to be shown in the output file. If you change this to echo = FALSE, then the code will be hidden and only the output of the code chunks will be seen in the output file.
There are also many other options that you can change, but we won’t go into those details in this workshop.
Text
Finally, you can include text in your R Markdown file. This is any text or explanation you want to include, and it’s formatted with Markdown. We’ll learn more about Markdown formatting soon!
Starting the report
Let’s return to the new R Markdown file you created and delete everything below the setup code chunk. (That stuff is just examples and reminders of how to use R Markdown.)
Next, let’s save our R markdown file to the reports directory.
You can do this by clicking the save icon in the top left or using control + s (command + s on a Mac).
There’s one other thing that we need to do before we get started with our report.
To render our documents into html format, we can “knit” them in R Studio.
Usually, R Markdown renders documents from the directory where the document is saved (the location of the .Rmd file), but we want it to render from the main project directory where our .Rproj file is.
This is because that’s where all of our relative paths are from and it’s good practice to have all of your relative paths from the main project directory.
To change this default, click on the down arrow next to the “Knit” button at the top left of R Studio, go to “Knit Directory” and click “Project Directory”.
Now it will assume all of your relative paths for reading and writing files are from the un-report directory, rather than the reports directory.
Now that we have that set up, let’s start on the report!
We’re going to use the code you generated yesterday to plot GDP vs. Life Expectancy to include in the report. Recall that we needed a couple R packages to generate these plots. We can create a new code chunk to load the needed packages. You could also include this in the previous setup chunck, it’s up to your personal preference.
```{r packages}
library(tidyverse)
```
Now, in a real report this is when we would type out the background and purpose of our analysis to provide context to our readers. However, since writing is not a focus of this workshop we will avoid lengthy prose and stick to short descriptions. You can copy the following text into your own report below the package code chunk.
This report was prepared to the attention of the United Nations. It analyzes the relationship between a country's GDP, its life expectancy and CO2 emissions. Our goal is to determine to what degree a country’s economic strength or weakness may be related to its public health status and impact on climate pollution. We hypothesize that both life expectancy and CO2 emissions increase with a country's GDP.
Now, since we want to show our results comparing GDP and life expectancy by country, we need to read in this data so we can regenerate our plot. We will add another code chunk to prepare the data.
```{r data}
gapminder_1997 <- read_csv("data/gapminder_1997.csv")
```
Now that we have the data, we need to produce the plot. Let’s create it!
```{r gdp_lifeexp_1997}
ggplot(data = gapminder_1997) +
aes(x = gdpPercap, y = lifeExp, color=continent, size=pop/1000000) +
geom_point() +
labs(x = "GDP Per Capita", y = "Life Expectancy",
title= "Do people in wealthy countries live longer?", size="Population (in millions)")
```
Now we can knit our document to see how our report looks! Use the knit button in the top left of the screen.
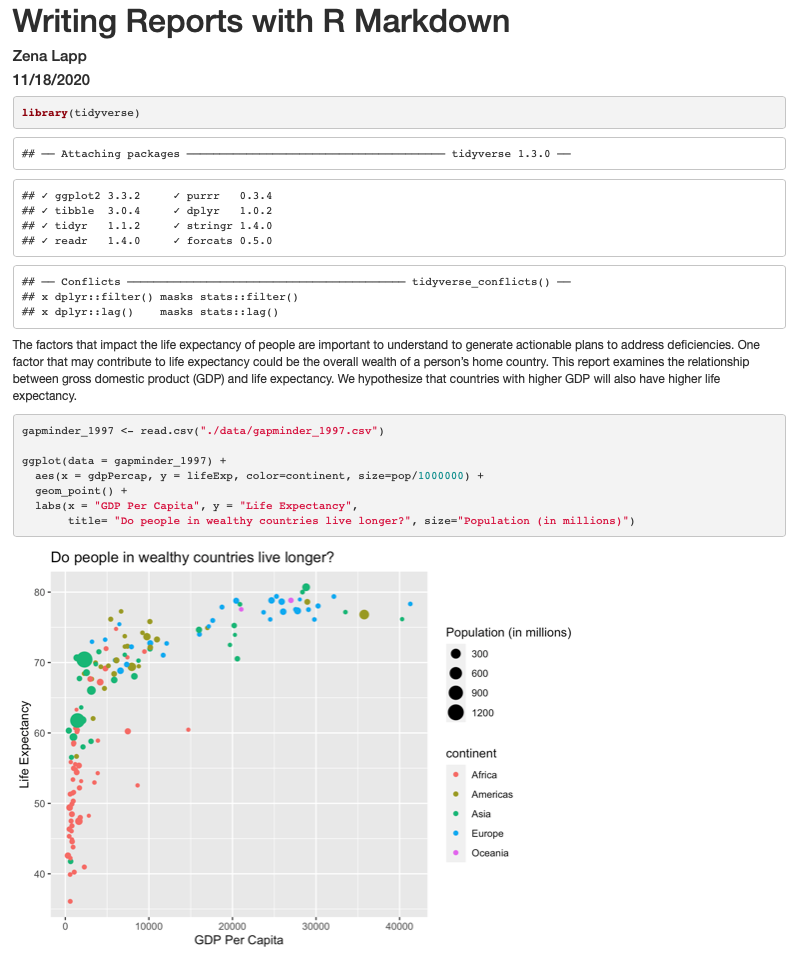
Amazing! We’ve created a report! Let’s push this to GitHub to make sure we preserve this document.
Now, all we need to do is commit our new report to git version control and update our remote repository using Git.
Returning to the command line,
First, we will check the status of our repository with the git status. This will tell us which files just changed.
Next, we will add the new files that we want tracked by git up with git add <filename(s)>. This adds any file(s) we list to version control.
Then we commit the file or changes with git commit -m "<your commit message>" and we add a message with the commit to describe the changes you are committing and it adds them to the git log.
Now, we can push those changes from our local computer to our github repo with git push. This will upload the changes to the git directory on the remote server.
If you don’t remember how to do this, you can review the git lesson.
It’s looking pretty good, but there seem to be a few extra bits that we don’t need in the report. For example, the report shows that we load the tidyverse package and the accompanying messages.
To get rid of this, we can revise our packages code chunk by adding include=FALSE just like in the setup chunk to prevent code and messages in this chunk from showing up in our report.
```{r packages, include=FALSE}
library(tidyverse)
```
We can also see the code that was used to generate the plot. Depending on the purpose and audience for your report, you may want to include the code. If you don’t want the code to appear, how can you prevent it? What happens if we add include=FALSE to the plot code chunk, too? Try rendering the R Markdown report with this change.

Oops! Now the plot doesn’t show up in our report at all. This is because setting include=FALSE prevents anything in the code chunk from appearing in the report. Instead we can add echo=FALSE to tell this code chunk that we don’t want to see the code but just the output.
```{r gdp_lifeexp_1997, echo=FALSE}
ggplot(data = gapminder_1997) +
aes(x = gdpPercap, y = lifeExp, color=continent, size=pop/1000000) +
geom_point() +
labs(x = "GDP Per Capita", y = "Life Expectancy",
title= "Do people in wealthy countries live longer?", size="Population (in millions)")
```
When we knit this again, our plot is back!
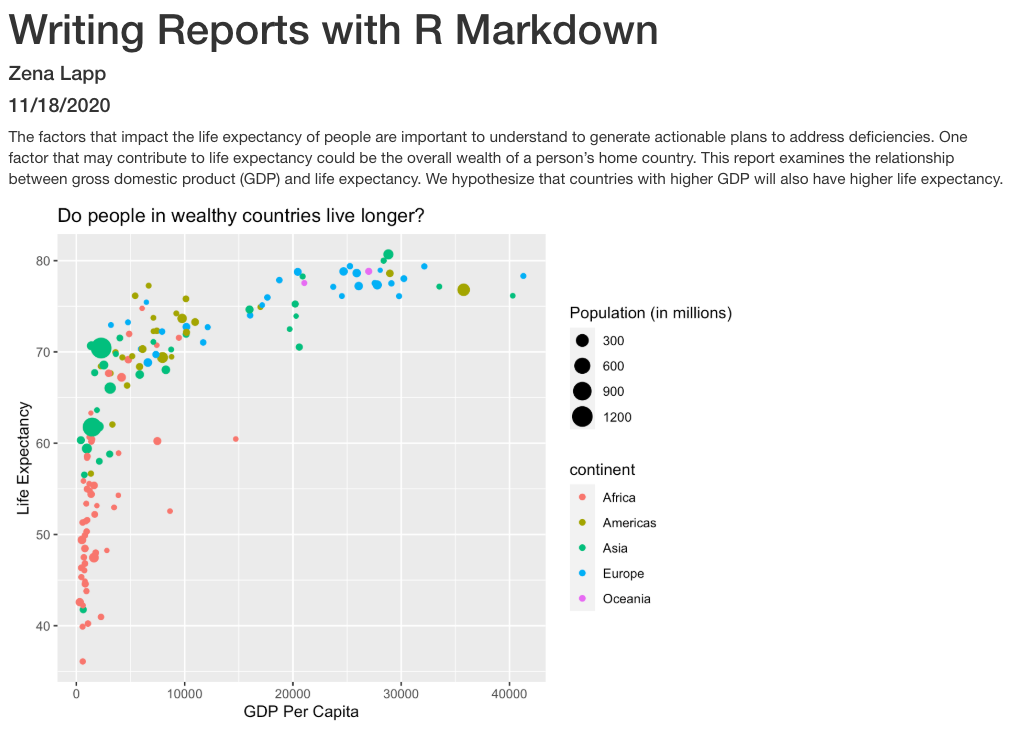
Before we finalize our report, let’s look at a few other cool features. Sometimes, you want to describe your data or results (like our plot) to the audience in text but the data and results may still change as you work things out. R Markdown offers an easy way to do this dynamically, so that the text updates as your data or results change. Here is how to do this.
First, let’s create a code chunk that summarizes features of our data that we can use to describe our plot to our audience. Note that we set include=FALSE because we only want this step to happen in the background. For our purposes, we will calculate how many countries were included in the analysis, as well as the minimum and maximum GDP per capita values.
Now, all we need to do is reference the values we just computed to describe our
plot. To do this, we enclose each value in one set of backticks
(`r some_R_variable_name `), while the r part once again
indicates that it’s a chunk of R code. When we knit our report, R will
automatically fill in the values we just created in the above code chunk. Note
that R will automatically update these values every time our data might change
(if we were to decide to drop or add countries to this analysis, for example).
The above plot shows the relationship between GDP per capita and life expectancy
for a total of `r nCountries ` countries. For this set of countries,
economic wealth ranged from a minimum of USD `r minGDP`
to a maximum of USD `r maxGDP` per capita.
In addition to reporting specific values in the text, we may also want to show a table of values. With R Markdown there are multiple ways to product tables. One way to generate smaller tables is manually. Using a special format we can generate a table in our output. Note that this does not get generated in a code chunk because it is markdown formatting not R code.
|HEADER 1|HEADER 2|
|-------------|-------------|
|row 1, column1|row 1, column 2|
|row 2, column1|row 2, column 2|
Columns are separated by the pipe key | located above Enter on the keyboard. The dashes distinguish the header row from the rest of the table. This header could be a name for each column or a Header for the whole table. Now that we know the basic structure we can fill in our table. This is how we could present the same numbers from the previous paragraph as a table instead, again using in-line code.
When we knit the report again, the code above will render like this:

|Summary of Data|
|------|------|
|Number of Countries|142|
|Minimum GDP per capita|312|
|Maximum GDP per capita|4.1283 × 10<sup>4</sup>|
This will render like this:

This is useful if we are reporting a few values, but can get tedious for larger tables. Another way we can add tables to our reports is using an R function called kable(). Since this is an R function, we will use it within a code chunk. We can give the kable() function a data table and it will format it to a nice looking table in the report. For example, we could use the following code to generate a table of all the countries in Oceania.
| country | pop | continent | lifeExp | gdpPercap |
|---|---|---|---|---|
| Australia | 18565243 | Oceania | 78.83 | 26997.94 |
| New Zealand | 3676187 | Oceania | 77.55 | 21050.41 |
Which will render like this:

Now that we have a report we are happy with, let’s push the changes to GitHub.
Formatting
We now know how to create a report with R Markdown. Maybe we also want to format the report a little bit to structure our thought process in a useful way (e.g., sections) and make it visually appealing? Markdown is a very simple programming language when it comes to syntax. Let’s try to figure out some syntax together. Suppose we wanted to create sections in our report.
Exercise: R Markdown headers
Try googling how to create sections by using headers and subheaders using R Markdown. What do you find?
Solution
We can easily create headers and subheaders by using the
#pound/hash sign. Our main headers have one#(e.g.# Main Header Here) and to create subheaders we add additinal#s (e.g.## First subheaderand### Second subheader)
OK, now that we know how to make headers, let’s practice some more Markdown syntax.
Exercise: R Markdown syntax
Go ahead and do some online searches on how to do the following:
- create a bullet point list with three items
- as the first item, write the name of your currently favorite programming language in bold
- as the second item, write the name of a function you have so far found most useful in italics
- as the third item, write one thing you want to learn next on your programming journey in bold and italics
- turn your bullet point list into a numbered list
- create a fourth list item and find an online guide and/or cheat sheet for basic Markdown syntax, write its name down here and hyperlink its url
Solution
This link has some helpful basic R Markdown syntax.
Integrating it all together: Paired exercise
You’ve learned so much in the past two days - how to use the Unix Shell to move around your computer, how to use git for version control and GitHub for collaborating with others on code, how to make pretty plots and do data analysis in R, and how to incorporate it all into a report. Now, you’re going to work in pairs to practice everything you learned. Ideally, you’ll have the same pair as for the git/GitHub lesson. Don’t worry - if you have questions, the instructor and helpers are here to help you out!
Only one of the people in your pair is going to create the R Markdown file. The other person is going to collaborate with that person using GitHub. So the first step is to choose one person in your pair to create/host the R Markdown file.
For the person who is going to host the new R Markdown file:
- Make a new R Markdown file.
- Give it an informative title.
- Delete all the unnecessary Markdown and R code (everything below the setup R chunk).
- Save it to the
reportsdirectory using an informative file name.
For the person who is going to collaborate with the host of the R Markdown file:
If you don’t already have your partner’s GitHub repo cloned from the git/GitHub lesson, clone their repo to your Desktop under the name USERNAME-un-report. If you don’t remember how to do this, you can review the git lesson.
The way you will collaborate with each other is as follows:
- For each exercise, both people will be thinking about how to answer the question, but only one person will be writing the code. This is called paired programming.
- Once you have completed an exercise, the person working on the exercise will add, commit, and push the changes to GitHub.
- Then the other person will pull the changes from GitHub.
- The person who pulled changes will code for the next exercise.
- Repeat the process for as many exercises as you can finish in the remaining time. Don’t worry if you don’t finish all of the exercises, and it’s not a race between groups! This is just a way for you to practice what you’ve learned.
One note: It may be helpful to copy and paste the questions into the R Markdown file as you go.
Exercises using the gapminder data
First we’re going to start out with a few questions about the gapminder dataset.
1. The very first step is to read in the gapminder dataset, so don’t forget! Also load the tidyverse package.
Solution
suppressMessages(library(tidyverse)) gapminder <- read_csv('data/gapminder_data.csv', show_col_types=FALSE) spec(gapminder)cols( country = col_character(), year = col_double(), pop = col_double(), continent = col_character(), lifeExp = col_double(), gdpPercap = col_double() )
Investigating population over time.
2. Make a scatter plot of year vs. population, separated into a plot for each contient. HINT: you can use facet_wrap(vars(column_name)) to separate into different plots based on that column.
Solution
ggplot(gapminder, aes(x=year,y=pop)) + geom_point() + facet_wrap(vars(continent))
3. It seems like there are 2 outliers - which countries are those?
Solution
gapminder %>% filter(pop > 1e9) %>% select(country) %>% unique()# A tibble: 2 × 1 country <chr> 1 China 2 India
4. Plot year vs. population separated into a plot for each continent but excluding the 2 outlier countries.
Solution
gapminder %>% filter(country != 'China' & country != 'India') %>% ggplot(aes(x=year,y=pop)) + geom_point() + facet_wrap(vars(continent))
Bonus questions: come back to these if you have time at the end
5. In the plot above, the years look kind of messy. Can you rotate the x axis text 90 degrees so that the years are more readable? Feel free to search the internet if you don’t know how to do this!
Solution
gapminder %>% filter(country != 'China' & country != 'India') %>% ggplot(aes(x=year,y=pop)) + geom_point() + facet_wrap(vars(continent)) + theme(axis.text.x=element_text(angle=90))
6. It’s hard to see which country is which here. Can you change the scatter plot to a line plot so we can get a better sense of trends over time? HINT: This website has more information: https://www.r-graph-gallery.com/line-chart-several-groups-ggplot2.html
Solution
gapminder %>% filter(country != 'China' & country != 'India') %>% ggplot(aes(x=year,y=pop,group=country)) + geom_line() + facet_wrap(vars(continent)) + theme(axis.text.x=element_text(angle=90))
Looking into life expectancy a bit more.
7. What country had the highest life expectancy in 1982? Hint: use the slice_max() function to get the row for a maximum value in a dataset. You can use ?slice_max and/or the internet to learn more about how to use the function.
Solution
# A tibble: 1 × 6 country year pop continent lifeExp gdpPercap <chr> <dbl> <dbl> <chr> <dbl> <dbl> 1 Japan 1982 118454974 Asia 77.1 19384.
8. Now, do the same thing but for all years! HINT: Use the group_by() function.
Solution
gapminder %>% group_by(year) %>% select(country,year,lifeExp) %>% slice_max(lifeExp) %>% print(n=Inf)# A tibble: 12 × 3 # Groups: year [12] country year lifeExp <chr> <dbl> <dbl> 1 Norway 1952 72.7 2 Iceland 1957 73.5 3 Iceland 1962 73.7 4 Sweden 1967 74.2 5 Sweden 1972 74.7 6 Iceland 1977 76.1 7 Japan 1982 77.1 8 Japan 1987 78.7 9 Japan 1992 79.4 10 Japan 1997 80.7 11 Japan 2002 82 12 Japan 2007 82.6
9. Make a boxplot for the life expectancies of the countries in Asia for each year (year is the x axis, life expectancy is the y axis). Also fix the x and y axis labels.
Solution
Bonus questions: come back to these if you have time at the end
10. What are the outliers in life expectancy in Asia for each year (lower life expectancy)?
Solution
gapminder %>% filter(continent == 'Asia') %>% group_by(year) %>% slice_min(lifeExp)# A tibble: 12 × 6 # Groups: year [12] country year pop continent lifeExp gdpPercap <chr> <dbl> <dbl> <chr> <dbl> <dbl> 1 Afghanistan 1952 8425333 Asia 28.8 779. 2 Afghanistan 1957 9240934 Asia 30.3 821. 3 Afghanistan 1962 10267083 Asia 32.0 853. 4 Afghanistan 1967 11537966 Asia 34.0 836. 5 Afghanistan 1972 13079460 Asia 36.1 740. 6 Cambodia 1977 6978607 Asia 31.2 525. 7 Afghanistan 1982 12881816 Asia 39.9 978. 8 Afghanistan 1987 13867957 Asia 40.8 852. 9 Afghanistan 1992 16317921 Asia 41.7 649. 10 Afghanistan 1997 22227415 Asia 41.8 635. 11 Afghanistan 2002 25268405 Asia 42.1 727. 12 Afghanistan 2007 31889923 Asia 43.8 975.
11. Make a boxplot for the life expectancies of the countries over time for each continent. Try to fix the x and y axis labels and text, too. Feel free to change the theme if you’d like.
Solution
12. Which country has had the greatest increase in life expectancy from 1952 to 2007? HINT: You might want to use the pivot_wider() function to get your data in a format with columns for: country, 1952 life expectancy, 2007 life expectancy, and the difference between 2007 and 1992 life expectancy.
Solution
# A tibble: 1 × 4 country `1952` `2007` diff <chr> <dbl> <dbl> <dbl> 1 Oman 37.6 75.6 38.1
13. What countries had a decrease in life expectancy from 1952 to 2007?
Solution
# A tibble: 2 × 4 country `1952` `2007` diff <chr> <dbl> <dbl> <dbl> 1 Swaziland 41.4 39.6 -1.79 2 Zimbabwe 48.5 43.5 -4.96
Exercises integrating a new dataset
If you finished the questions involving the gapminder dataset (bonus questions are optional), move on to these questions next. Note that we don’t expect you to finish all of these! You can also use them as practice after the workshop if you’d like.
Now that you’ve practiced what you’ve learned with the gapminder data, you’re going to try using what we’ve learned to explore a new dataset.
Preview of the data
This dataset has information on the gross domestic expenditure on research and development (R&D) for different countries. We’re going to use it to practice the data analysis workflow that you learned over the course of the workshop.
Data: Gross domestic expenditure on research and development (R & D)
Data source: UN data, under “Science and technology”
Data path: data/SYB63_286_202009_GDP_on_RnD.csv
Raw csv file:
T27,Gross domestic expenditure on research and development (R&D),,,,,
Region/Country/Area,,Year,Series,Value,Footnotes,Source
1,"Total, all countries or areas",2005,Gross domestic expenditure on R & D: as a percentage of GDP (%),1.5261,,"United Nations Educational, Scientific and Cultural Organization (UNESCO), Montreal, the UNESCO Institute for Statistics (UIS) statistics database, last accessed June 2020."
1,"Total, all countries or areas",2010,Gross domestic expenditure on R & D: as a percentage of GDP (%),1.6189,,"United Nations Educational, Scientific and Cultural Organization (UNESCO), Montreal, the UNESCO Institute for Statistics (UIS) statistics database, last accessed June 2020."
...
32,Argentina,2017,Gross domestic expenditure on R & D: Business enterprises (%),16.5161,,"United Nations Educational, Scientific and Cultural Organization (UNESCO), Montreal, the UNESCO Institute for Statistics (UIS) statistics database, last accessed June 2020."
...
CO2 dataset (UN data) preview:
T24,CO2 emission estimates,,,,,
Region/Country/Area,,Year,Series,Value,Footnotes,Source
8,Albania,1975,Emissions (thousand metric tons of carbon dioxide),4338.3340,,"International Energy Agency, IEA World Energy Balances 2019 and 2006 IPCC Guidelines for Greenhouse Gas Inventories, last accessed June 2020."
8,Albania,1985,Emissions (thousand metric tons of carbon dioxide),6929.9260,,"International Energy Agency, IEA World Energy Balances 2019 and 2006 IPCC Guidelines for Greenhouse Gas Inventories, last accessed June 2020."
Reading in and cleaning the data
1. First, read in the data. Note that you need to skip the first line of the file because that’s just a title for the whole dataset (see above). Also rename the columns to something more informative (as you learned, there are lots of ways to do this, and different preferences - feel free to use whichever method you want!).
Solution
rnd <- read_csv('data/rnd-un-data.csv', skip = 1) %>% rename(country=`...2`) %>% rename_all(tolower)New names: * `` -> ...2Rows: 2420 Columns: 7── Column specification ────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────── Delimiter: "," chr (4): ...2, Series, Footnotes, Source dbl (3): Region/Country/Area, Year, Valueℹ Use `spec()` to retrieve the full column specification for this data. ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
2. Next, take a look at the series column (or whatever you renamed it to), and make the titles shorter and with no spaces to make them easier to work with.
Solution
rnd <- rnd %>% mutate(series=recode(series, "Gross domestic expenditure on R & D: as a percentage of GDP (%)" = 'gdp_pct', "Gross domestic expenditure on R & D: Business enterprises (%)" = 'business', "Gross domestic expenditure on R & D: Government (%)" = 'government', "Gross domestic expenditure on R & D: Higher education (%)" = 'higher_ed', "Gross domestic expenditure on R & D: Funds from abroad (%)" = 'abroad', "Gross domestic expenditure on R & D: Not distributed (%)" = 'not_distributed', "Gross domestic expenditure on R & D: Private non-profit (%)" = 'non_profit'))
3. Next, make a column for each of the data types in the series column (or whatever you renamed it to). This should give you the following columns: country name, year, expenditure in general, % of funds from business, % of funds from government, % of funds from higher ed, % of funds from non-profit, % of funds from abroad, % of funds from non-specified sources.
Solution
rnd <- rnd %>% pivot_wider(names_from=series,values_from=value)
Note that there is a lot of missing data.
Now we have our data set up in a way that makes it easier to work with. Feel free to clean up the data more before moving on to the next step if you’d like.
Plotting with the R & D dataset
1. Plot the distribution of percent expenditure using a histogram. Observe the range and how heavy the “tails” are. Note: You will likely get a note and a warning. Feel free to change the number of bins as the note describes. Why do you get warnings? (Hint: Look at the column you’re plotting.). Bonus: Plot the same exact data in a way in which you don’t get warnings.
Solution
rnd %>% filter(!is.na(gdp_pct)) %>% ggplot(aes(x=gdp_pct)) + geom_histogram(bins=30)
2. Plot the expenditure by year (discrete x vs continuous y) using a scatter plot. Feel free to try to make the plot more legible if you want.
Solution
rnd %>% filter(!is.na(gdp_pct)) %>% ggplot(aes(x=as.character(year), y=gdp_pct)) + geom_point(alpha=0.5) + xlab('')
3. Plot the expenditure by year (discrete x vs continuous y) using a violin plot or a boxplot.
Solution
rnd %>% filter(!is.na(gdp_pct)) %>% ggplot(aes(x=as.character(year), y=gdp_pct)) + geom_violin() + xlab('')
Combining the CO2 and R&D datasets
Now we’re going to work with the CO2 and R&D datasets together.
Unfortunately, we don’t have the exact same dates for all of them.
1. First, read in the CO2 dataset. You can use the code from the R for data analysis lesson to clean the CO2 data.
Solution
# read in and clean CO2 data co2 <- read_csv("data/co2-un-data.csv", skip=2, col_names=c("region", "country", "year", "series", "value", "footnotes", "source"), show_col_types = FALSE) %>% select(country, year, series, value) %>% mutate(series = recode(series, "Emissions (thousand metric tons of carbon dioxide)" = "total", "Emissions per capita (metric tons of carbon dioxide)" = "per_capita")) %>% pivot_wider(names_from=series, values_from=value) spec(co2)NULL
2. Merge the CO2 dataset and the R&D dataset together. Keep only the following colums: country, year, total CO2 emissions, CO2 emissions per capita, and percent of GDP used for R&D.
Solution
co2_rnd <- full_join(co2, rnd) %>% select(country, year, total, per_capita, gdp_pct)Joining, by = c("country", "year")
3. You might have noticed that we don’t have both CO2 data and R&D data for all years. Filter the merged dataset so that you only keep country/year combinations that have both C02 and R&D data (i.e., only keep rows for which CO2 and R&D values are not missing). HINT: Search the internet for the use of is.na() and ! (the “not operator”) to help you here.
Solution
co2_rnd <- co2_rnd %>% filter(!is.na(total) & !is.na(gdp_pct))
4. How many countries per year do you have after filtering? HINT: You can use summarize(count=n()) to help you out.
Solution
co2_rnd %>% group_by(year) %>% summarize(count=n())# A tibble: 5 × 2 year count <dbl> <int> 1 2005 83 2 2010 86 3 2015 94 4 2016 11 5 2017 57
Plotting with the CO2 and R&D datasets together
1. Plot R&D expenditure vs. CO2 emission for each country using a scatter plot. You can choose total or per-capita CO2 emissions, or both. Make sure you have informative x and y axis labels.
Solution
ggplot(co2_rnd, aes(x=gdp_pct,y=per_capita)) + geom_point() + ylab('Per-capita CO2 emissiosn') + xlab('Percent of GDP spent on R&D')
2. Next, facet the above plot by year.
Solution
ggplot(co2_rnd, aes(x=gdp_pct,y=per_capita)) + geom_point() + ylab('Per-capita CO2 emissiosn') + xlab('Percent of GDP spent on R&D') + facet_wrap(vars(year))
3. Identify the countries that have five time points for both C02 emissions and R&D.
Solution
co2_rnd %>% group_by(country) %>% summarize(count=n()) %>% filter(count==5)# A tibble: 3 × 2 country count <chr> <int> 1 Azerbaijan 5 2 Cuba 5 3 Panama 5
BONUS
4. For three of the countries you identified, plot the Percent of GDP spent on R&D and the per-capita CO2 emissions over time on the same plot. Color the two different values differently. HINTS: Use pivot_longer to get the data in the right format. Search the internet for how you can use %in%. Also remember the c() function from earlier in the workshop that can be used to combine values into a vector or list.
Solution
co2_rnd %>% filter(country %in% c('Azerbaijan','Cuba','Panama')) %>% pivot_longer(c(per_capita,gdp_pct)) %>% ggplot(aes(x=year,y=value,col=name)) + geom_line() + facet_wrap(vars(country))+ scale_color_discrete(name = "", labels = c("GDP %", "CO2 Per Capita"))
Bonus questions
5. For the R&D dataset, each country can have data for one or multiple years. What is the range of numbers of yearly data points for each country, and how many countries are there for each value within the range? (e.g., x countries have two different years and y have five years)
6. After merging the data sets, there is some missing data. How many NAs are present in each data column for the R&D data set? How may these missing data affect our intuitive observation in a plot and/or summary statistic? (e.g., ggplot removes NAs but stat functions (e.g., median()) often ask for specific input regarding how to deal with NAs).
7. What countries have the highest and lowest data availabilities? (How would you measure data availability?)
8. Add in the gapminder dataset and color the scatter plots we made above by continent, also facet by continent.
9. For a specific continent (e.g. Europe), show the importance of business and government funding for R&D. Do this using a stacked bar plot.
10. Create an R Markdown report with some of the information from these exercises. Decide exactly what you want to focus your report on, and then also perform additional analyses to include in your report. Also make sure your figures are legible and understandable!
Key Points
R Markdown is an easy way to create a report that integrates text, code, and figures.
Options such as
includeandechodetermine what parts of an R code chunk are included in the R Markdown report.R Markdown can render HTML, PDF, and Microsoft Word outputs.
Conclusion
Overview
Teaching: 15 min
Exercises: minQuestions
What do I do after the workshop to apply what I learned and keep learning more?
Where can I learn more coding skills?
How do I deal with coding errors (i.e. debug)?
What resources are there at the University of Michigan?
What other coding concepts should I learn?
Objectives
Learn how to get help with code via the Internet and at the University of Michigan.
Learn about other coding concepts that would be good to learn in the future.
Where to go from here?: Departing on your own coding journey
We have come to the end of this workshop. You learned some basic procedures for importing, managing, visualizing and reporting your data.
As you continue on your coding journey, two things will happen:
- You will encounter bugs and need to figure out how to solve them (“debugging”), and
- You will want to learn new data processing and analysis techniques.
As we complete the course, we want to share with you some tips and tricks that have helped us on our own programming journeys.
Writing code at the University of Michigan
There are many local opportunities at the University of Michigan or around the Ann Arbor campus to find coding support, learn new programming skills, and connect with other users.
Get help and connect
- Software Carpentry Workshops
- We’re always looking for more helpers & instructors!
- CoderSpaces, CoderSpaces Slack Space
- Data Analysis Networking Group (DANG!)
- CSCAR consulting and workshops
- Local meetup groups
Dealing with coding errors
Even well seasoned coders run into bugs all the time. Here are some strategies of how programmers try to deal with coding errors:
- Don’t panic. Bugs are a normal part of the coding process. Think of each bug as a great opportunity for becoming better at writing code. When you’re debugging, you’re being a detective and trying to figure out what’s wrong. Think of it as a fun mystery to solve! You’ll become a better detecive the more you practice.
- First, determine which type of problem you are having: either the code tries to run but returns an error message, or your code runs without error but you did not get the result you expect.
- If you are getting an error message, read the error message carefully. Unfortunately, not all error messages are well written and it may not be obvious at first what is wrong. Try to determine if you are getting a syntax error or a runtime error.
- A syntax error is when the program doesn’t understand how to run the code you provided. In R, these error messages usually contain the word “unexpected”. Often these are because of a typo. Check that your parentheses and quotes are balanced and check that you haven’t misspelled a variable or function name. It’s difficult to identify the exact location where an error starts so you may have to look at lines before the line where the error was reported. In RStudio, look at the code coloring to find anything that looks off. RStudio will also put a red x or an yellow exclamation point to the left of lines where there is a syntax error.
- A runtime error is when the program understood all the commands you wrote, but ran into an error when trying to run them. If you are running multiple commands, try running each on its own. Before each command, check that you are passing the values you expect; and after each command, verify that the results seem sensible.
- If you have been able to find parts of your code that cause the error, take some time to review the documentation for the functions or commands you are using. Even for functions or commands you’ve used before, there may still be parts of the documentation that you just haven’t needed to use before. See if there are examples in the documentation that you can compare your own code to. Most programs make finding the documentation pretty easy.
- R: Typing a question mark immediately followed by a function name, i.e.,
?<command>(e.g. ?print), pulls up the R documentation for that function. This provides a function description, as well as in-depth information about each function argument including its purpose, default and other available options. - Shell: Using
man <command>or<command> --helppulls up the manual page with more information. To quit, clickq. - Git:
git helpdisplays help information about git. Read more here.
- R: Typing a question mark immediately followed by a function name, i.e.,
- Search online for the error message along with the function that is not working. Most likely, someone encountered this error before you and the examples online will help you figure out your own issue.
- Perform a general online search for what you are trying to do, e.g., “how to import a comma separated spreadsheet into R”, “how to delete a directory from the command line”, “how to merge conflicts in git”. Typically, you will find step-by-step online documentation that you can adapt for your own purposes.
Debugging code
If searching for your particular code problem hasn’t turned up a solution, you may have to do a bit of debugging. Debugging is the process of finding exactly what caused your error, and changing only what is necessary to fix it. There are many strategies to debugging code. Consider checking out the following resources to learn more about it.
- “5 Essential Tips to Debug Any Piece of Code” by mayuko [video, 8min] - Good general advice for debugging.
- “Object of type ‘closure’ is not subsettable” by Jenny Bryan [video, 50min] - A great talk with R specific advice about dealing with errors as a data scientist.
Asking strangers for help
If you are unable to determine what’s wrong with your own code, the internet offers several possible ways to get help: asking questions on programming websites, interacting with developers on GitHub, chatting with other programmers on Slack, or reaching out on Twitter. If you’re intimidated by asking people on the internet, you can also reach out to people at the University of Michigan. You don’t have to do this all on your own. However, there are some important things to keep in mind when asking questions - whether it be to people on the internet, or to people at the university. You may want to consider these tips to help you increase your chances of getting the support you need:
- First, provide a brief summary of what you are trying to accomplish. You may have been working on the problem for a long time but the person you are asking will have no idea of your work until now. Keep it short and focus on your ultimate goal rather than just talking about your code. Sometimes there may better ways to solve the problem than the solution you’ve attempted this far.
- Then, briefly mention what you’ve tried so far. Maybe mention the search terms you used online or the specific documentation you’ve looked at. It should be clear that you’ve at least tried to solve the problem yourself first (after all, that’s the way you’re most likely to learn the most).
- Now, describe the problem you are having. Don’t just say “the code doesn’t work.” Are you getting an error? If so, what is the exact message? Are you getting unexpected results? If so, then what exactly were you expecting to happen?
- Next, include a minimal, reproducible example. This means you need to take away any parts of your code that aren’t necessary to create the error you are dealing with. Often, this may mean starting a branch with a new program just for testing. Make sure the code itself can be run by including or randomly generating some sample data. If there’s no way to copy/paste and run the code on a computer, that means the individuals you are asking for help need to run the code in their head which is a lot harder than running code on a computer. Clearly describe the desired behavior so that possible solutions can be tested and verified. In the process of creating a minimal, reproducible example, it is possible you will solve your own problem.
- Keep in mind the person helping you is unlikely to know the solution off the top of their head. Even the best programmers don’t have all the documentation memorized. But with time, programmers get better at solving problems based on their past experiences. By providing a reproducible example, you allow someone else to use their debugging skills to help you rather than making them guess what might be wrong.
- If it’s not obvious from your question, it can be very useful to include details about the operation system you are using and the versions of the programs you are currently running. Sometimes, things work differently as programs change over time and the code you found on the web may have been for a different version.
- Make it easy to reply. Try to ask one specific question. Asking a bunch of questions takes more time and energy to respond, and you want to minimize the work you create for the kind stranger who is helping you out. Also, if you include sample data with which to test possible solutions, then someone can verify their suggestion works before replying. This will help minimize back-and-forth with your helper which can take time.
- Finally, recognize that asking a good question is an art, not a science. It is something you will get better at with more time. You can also get better by trying to help other people with their questions. That way, you can see what information is necessary to really help someone. It’s a delicate balance between providing enough detail so it’s possible to intelligently respond yet keeping things short enough to not overwhelm your helper (more isn’t always better).
Learning new code
Free open-source programming languages such as bash, Git and R are constantly evolving. As you try out new data processing and analysis techniques, you will continue to learn new coding logic, concepts, functions, and libraries. Widely available user tools and documentation are a main benefit of free open-source software.
In the following, we list some strategies and resources we find useful. As you move forward, you are likely to identify other resources that fit your own learning style.
General
- Stack Overflow: Users post questions or errors and other users respond. (This is a great place to search for help but might not be the best place to ask a question if you are a beginner.)
- Cheat Sheets!! You can find many different cheat sheets for entire languages or specific libraries or even functions online. Just search for “
cheat sheet" to see what is available. We have listed a few below. - Study other people’s code. Increasingly, people publish their code (e.g., in a GitHub repository) to facilitate reproducible research. You can take someone else’s code as a starting point and adapt it to your own needs.
- Other people in your department or lab are also great resources for asking about specific coding problems.
R
- RStudio Community Forum. This is a site that is welcoming to new R users where you can ask R programming questions.
- Search R Package Documentation. This site is very useful when you see someone using a function but don’t know which package it comes from. This site lets you search across the help pages for all R packages.
- RStudio Cheat Sheets. A good collection of cheat sheets to print out and hang at your desk. For example: base R, tidyverse, and ggplot
- If there is a specific R package that you want to use (e.g. mikropml for machine learning analysis), you can use the vignettes to help you learn the basics of the package. You can search for the vignette online, or use the
vignette()function in R to open them.
R Markdown
Free learning platforms available at U-M
Some important advanced coding concepts that you will want to learn if you continue coding a lot
There are some coding concepts that we did not have time to cover in this workshop, but are important to learn as you continue on your journey and begin to perform more sophisticated data analysis projects. While we have not created resources for these topics, we provide some links to where you can learn more. Note that these are more advanced coding topics; you should be come comfortable with what you learned in the workshop before trying to delve deeper into these other concepts. However, you’ll likely come across situations where one of these will be useful, and that’s when you should learn it!
We’ve provided some links below, but feel free to search for other explanations and tutorials as well.
R coding topics
Some more advanced R coding topics include:
Here is a nice tutorial on conditionals, loops, and functions all together.
Domain-specific analyses
We encourage you to investigate domain-specific packages and software that will help you perform specific tasks related to your own research. The best way to find these packages is to either ask other people in your field and/or search for specific tasks that you would like to perform. If you’d like to perform the task in R, include that in your search (e.g. “find pairwise distances for DNA sequences in R” will help you find the R package ape which has a number of functions to perform phylogenetic and evolutionary analyses in R.)
High-performance computing clusters
If you’re performing computationally-intensive analyses, you’ll likely want to use a high-performance computing cluster. At the University of Michigan, many of us work on Great Lakes for much of our research. It can be a bit overwhelming at first, so try to find someone to help you learn the ropes. Sometimes there are also workshops where you can learn more.
Git/GitHub
If you start using Git/GitHub more frequently, it’s useful to learn how to create branches to work on new parts of your analysis. When you’re confident that it works, you can then merge the contents of the branch back into your “master” branch.
Key Points
When it comes to trying to figure out how to code something, and debugging, Internet searching is your best friend.
There are several resources at the University of Michigan that you can take advantage of if you need help with your code.
We didn’t have time to cover all important coding concepts in this workshop, so definitely continue trying to learn more once you get comfortable with the material we covered.
There are often packages and tools that you can leverage to perform domain-specific analyses, so search for them!