Introducing R and RStudio
Objectives
- Introduce R and RStudio.
- Learn how to read in a csv file.
- Discuss object naming conventions and types.
- Discuss functions and parameters.
- Learn how to get help with functions.
- Get comfortable with errors and asking for help.
The gapminder dataset
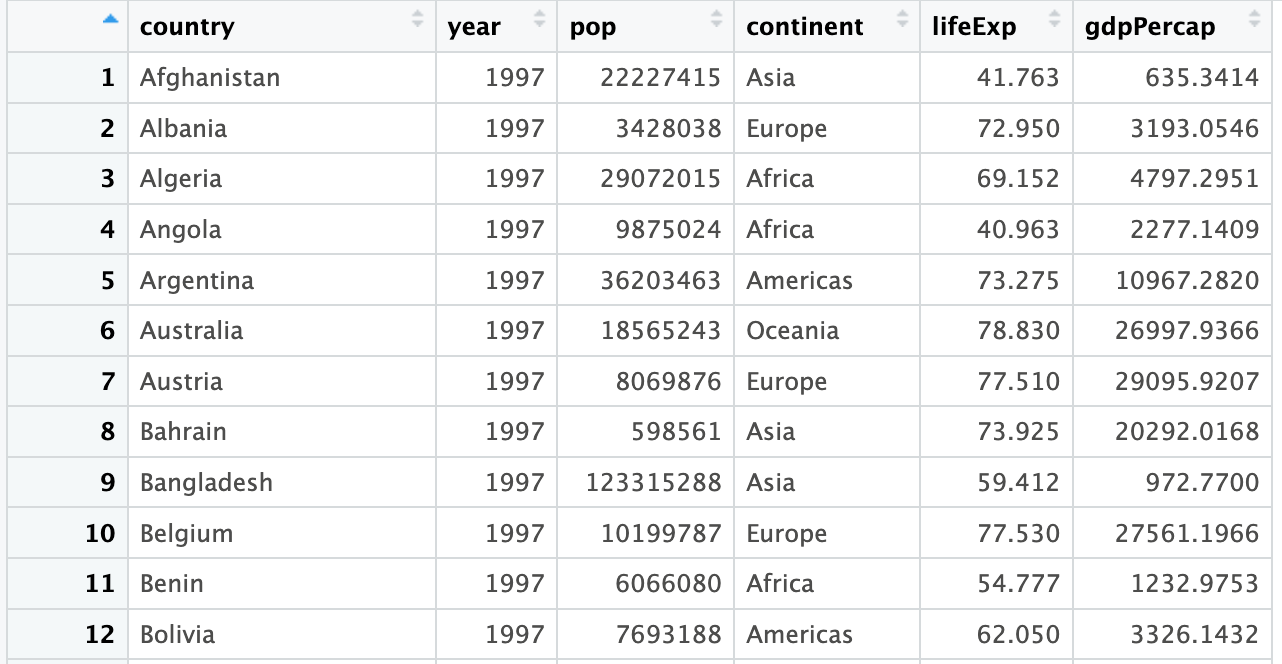
We will be using a dataset from gapminder which contains life expectancy, GDP, and population for countries around the world from 1952 to 2007. It is sufficiently rich to allow us to explore data manipulation and visualization in R. We’ll begin with data just from 1997, and will expand to the full 1952 - 2007 dataset towards the end. The following is a preview of the data we’ll be working with:
Server login
Let’s log in to the workshop server: http://bfx-workshop02.med.umich.edu
The login page for the server looks like:
Enter your user credentials and click Sign In. The RStudio interface should load and look like:
Checkpoint
Create a project
We will create an RStudio Project to easily keep track of our working directory. See the Projects section of R for Data Science for a more in-depth description of what a project is and how it’s helpful.
To create a Project, click File then New
Project…. In the New Project Wizard window that opens, select
Existing Directory, then Browse…. In the Choose
Directory window, select the IRR folder by clicking it
once, and then click the Choose button. Finally, click
Create Project.
Once we do this, RStudio will restart and the Files pane (lower
right) should put us in the ~/IRR folder where there is an
inputs/ folder and an IRR.Rproj file.
Checkpoint
The RStudio interface
RStudio is an integrated development environment where you can write, execute, and see the results of your code. The interface is arranged in different panes:
- The Console pane along the left where you can enter commands and execute them.
- The Environment pane in the upper right shows any variables you have created, along with their values.
- The pane in the lower right has a few functions:
- The Files tab let’s you navigate the file system.
- The Plots tab displays any plots from code run in the Console.
- The Help tab displays the documentation of functions.
Commands in the Console
Working directly in the console is working directly with R. Commands can be entered and run with the Enter key:
> 2+2
[1] 4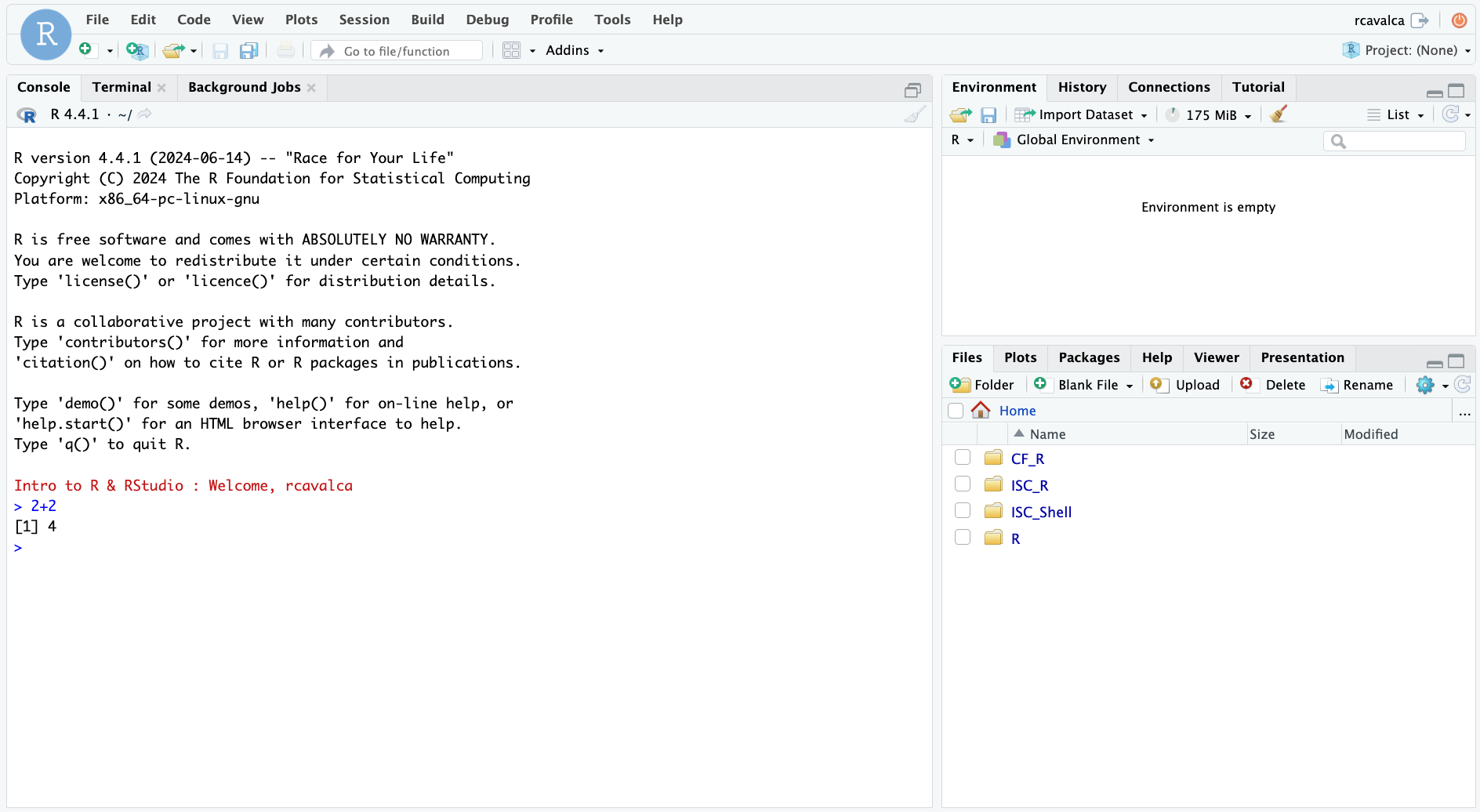
Checkpoint
Commands in a Script
Instead of entering commands directly into the Console, we’ll record them in and run them from a script. Some benefits to using a script rather than using the Console pane:
- Scripts record the steps taken to analyze the data.
- Scripts can be re-run, allowing for reproducibility.
- Scripts are can be shared.
We’ll create a script file by clicking on the icon in the upper-left of the interface (a blank piece of paper with a + sign), and selecting R Script.
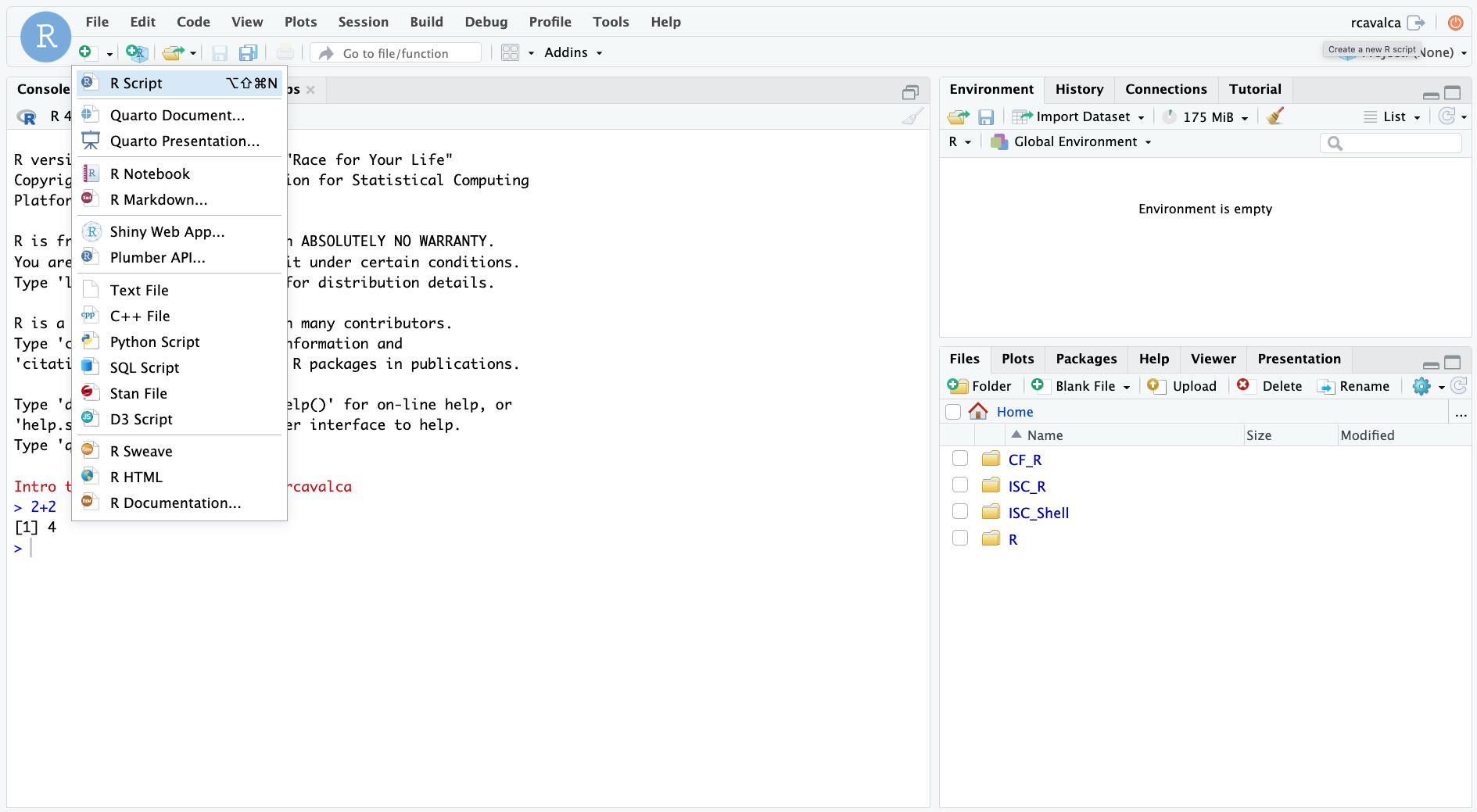
The new pane that opens is the Source pane, and you can think of it as a text editor:
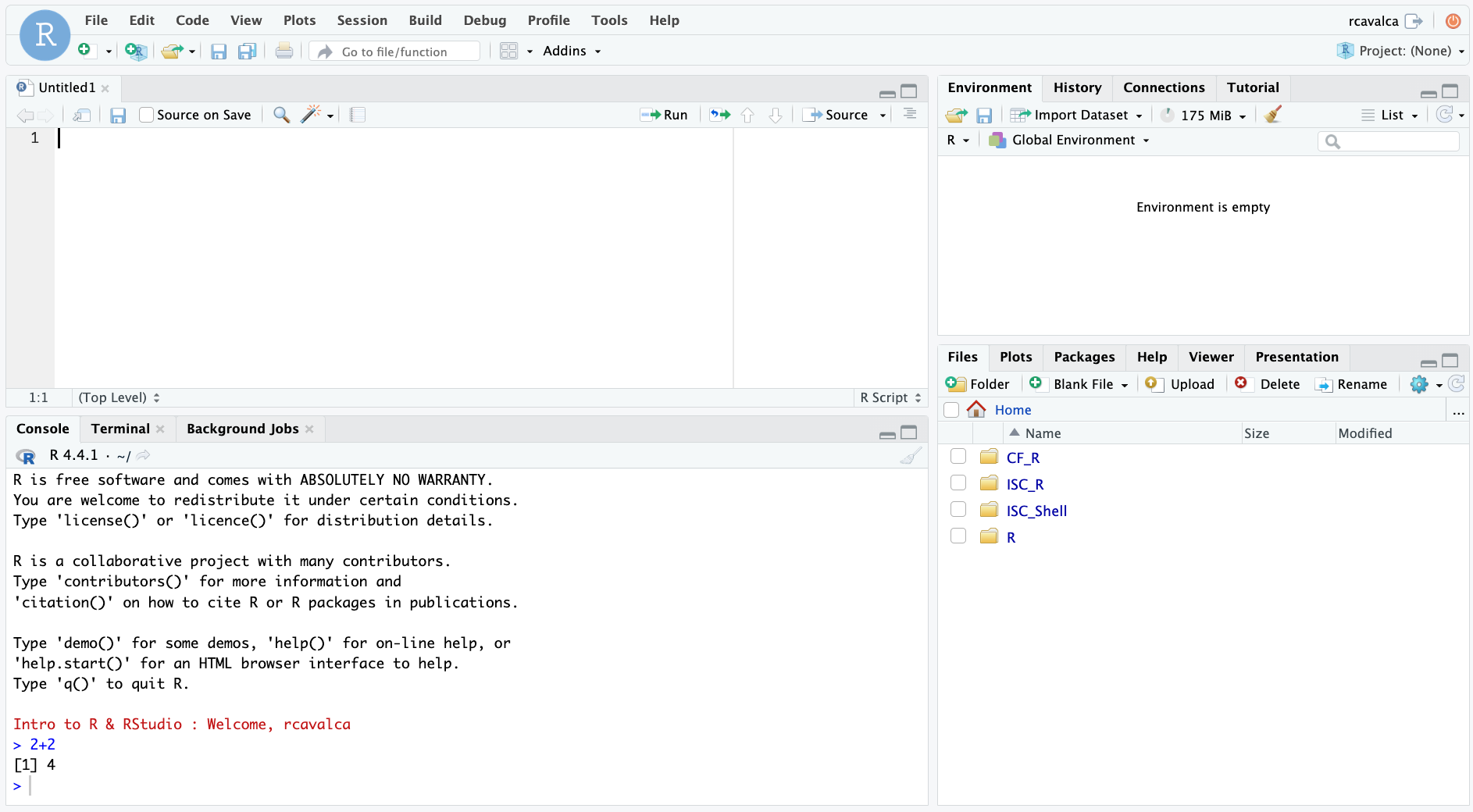
Code entered in a script file must explicitly be sent to the Console for execution with the Ctrl + Enter command. Enter the following command into the script file and execute it with Ctrl + Enter:
3+2Looking in the Console we see the executed line and its result. Note that pressing Enter in the script creates a new line and does not execute the code as in the Console.
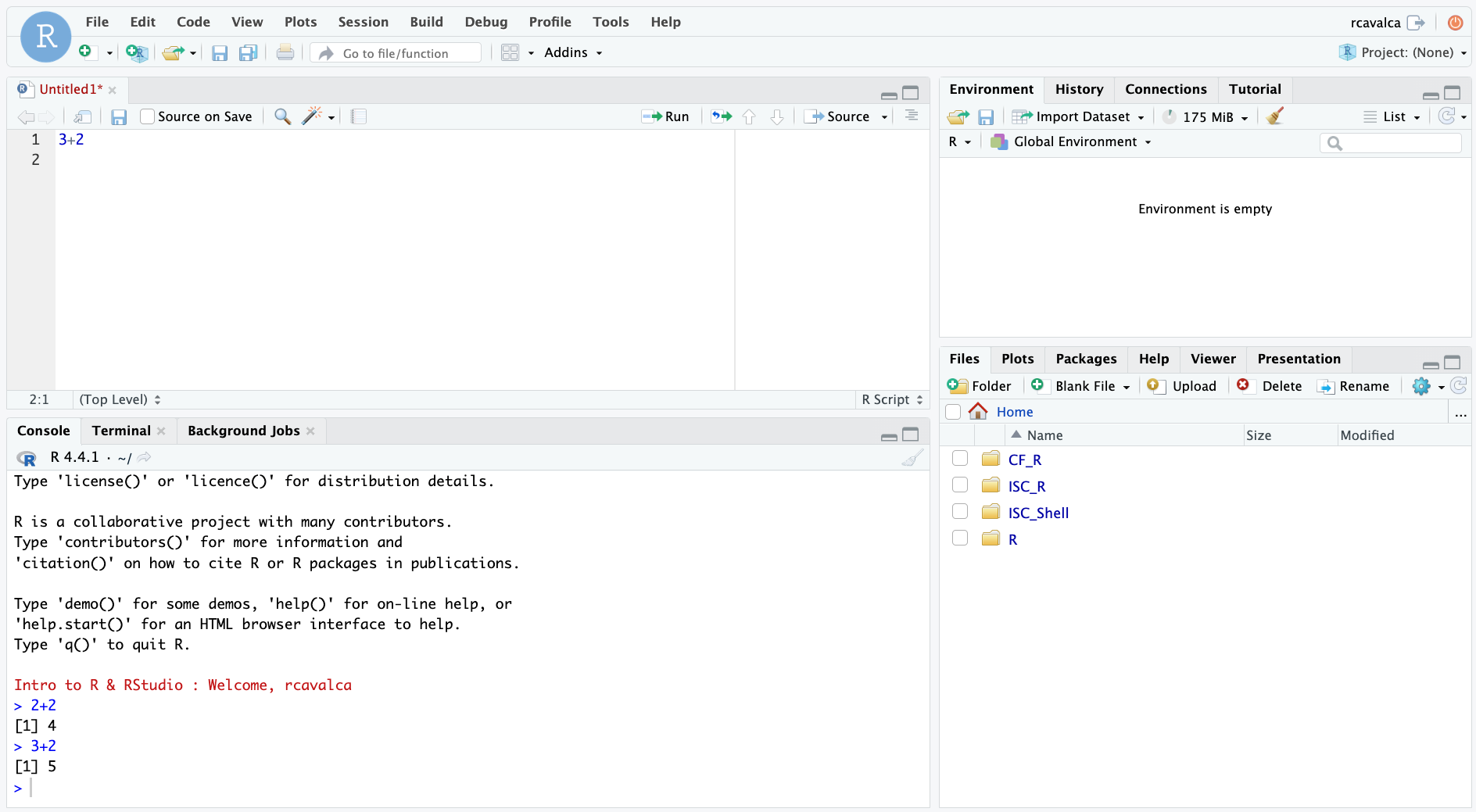
Console vs Script
The key differences between the Console and Script panes in RStudio:
| Console | Script |
|---|---|
| Reset across sessions | Preserved acros sessions |
| Run with Enter | Run with Ctrl + Enter |
| Inconvenient to share | Convenient to share |
Configuring RStudio
All of the panes in RStudio have configuration options. For example, you can minimize/maximize a pane or resize panes by dragging the borders. The most important customization options for pane layout are in the View menu. Other options such as font sizes, colors/themes, and more are in the Tools menu under Global Options.
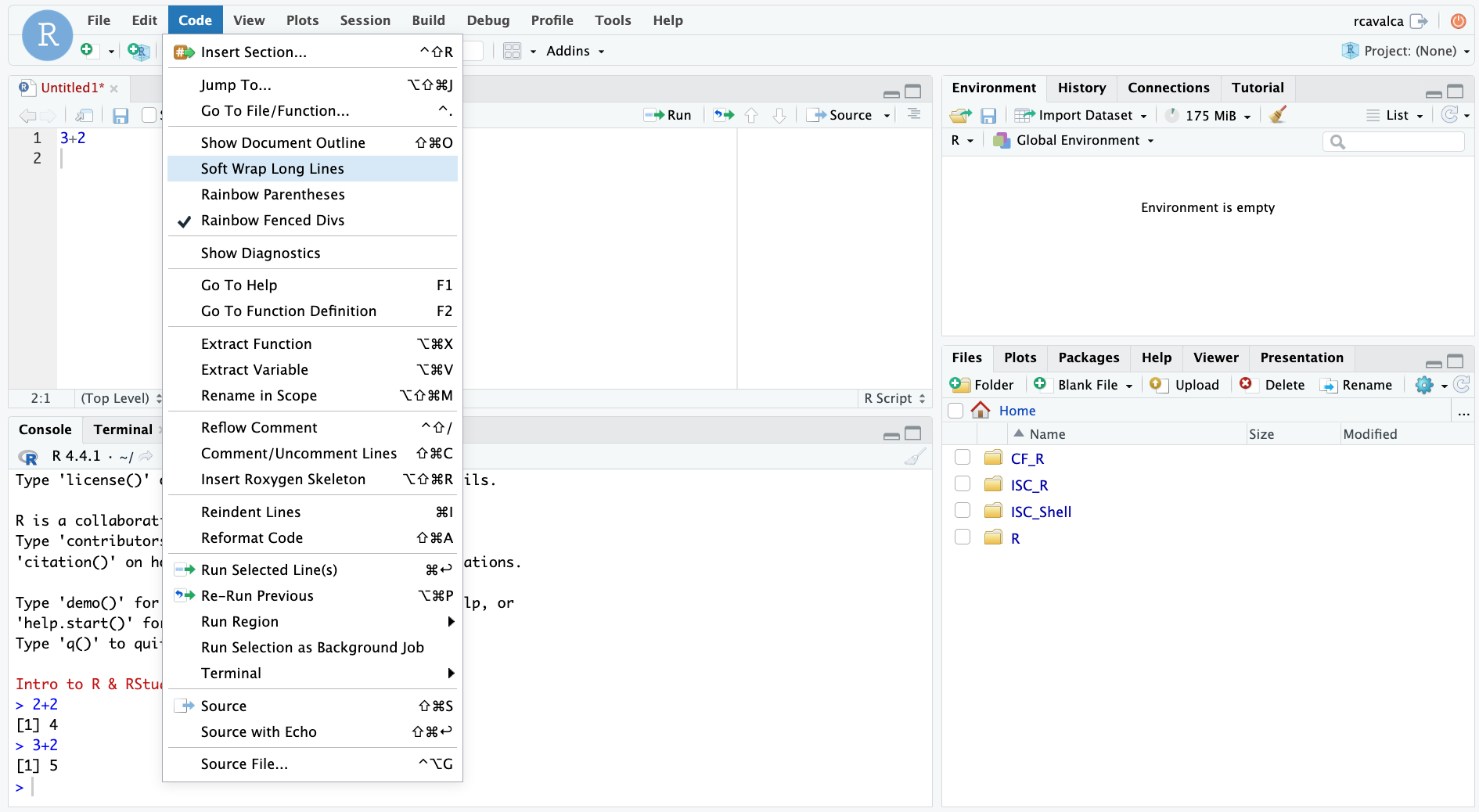
We can enable soft-wrapping of code by selecting Code and then Soft Wrap Long Lines.
Workshop flow
To accommodate different learning styles and to keep us moving along, we’ll provide code in three different ways, and you can get that code into RStudio in corresponding ways:
| Source of Code | Execution of Code |
|---|---|
| Zoom screen share | Type the code yourself. |
| Slack | Copy and paste into RStudio. |
| Website | Copy with code block button and paste into RStudio. |
Questions?
Directory structure
Before we begin, the folder structure of a project organizes all the relevant files. Typically we make directories for the following types of files:
- Raw data, called
data,input, etc, - Results, often
resultsoroutputwith subfolders fortables,figures, andrdata, and - Scripts, often
scripts.
We’ve already provided the raw data in the data/ folder,
and it’s generally a good idea to keep raw data in its own folder.
Let’s create some folders for our analysis scripts and results thereof.
# -------------------------------------------------------------------------
# Create directory structure
dir.create('scripts', recursive = TRUE, showWarnings = FALSE)
dir.create('results/figures', recursive = TRUE, showWarnings = FALSE)
dir.create('results/tables', recursive = TRUE, showWarnings = FALSE)
dir.create('results/rdata', recursive = TRUE, showWarnings = FALSE)Saving scripts
Let’s save our currently open script by clicking File and
then Save. Double click the scripts/ folder and
enter the file name ISC_day1.R.
Checkpoint
Getting started
Out of the box, R has a number of useful functions, but its power
lies in extending its functionality with packages / libraries. You can
think of libraries as collections of functions organized around a
particular functionality. For example, the tidyverse
package is a collection of packages that are designed to work together
to make data manipulation and visualization easier. In order to gain
access to functions in a package, we need to load the package into our R
session.
Loading Libraries
Let’s begin with loading tidyverse since that’s the
package we’ll use for the rest of the workshop.
# -------------------------------------------------------------------------
# Load the tidyverse package
library(tidyverse)Note: Package loading messages
Loading a package can result in a lot of feedback from R. These aren’t necessarily errors, but give more information about the result of loading the package. The output tells us which packages were loaded (note that
tidyverseis sort of a meta-package of packages). The first section of the output states which packages were lodaed and their versions. The second section notes “Conflicts” that occur because the name of a function is used multiple times. Sodplyr::filter() masks stats::filter()means that thedplyrlibrary and thestatslibrary have functions calledfilter(), and that when callingfilter(), thedplyrversion will be the default.
Checkpoint
Loading a csv file
Let’s jump right in and load some of the gapminder data using the
read_csv() function:
# -------------------------------------------------------------------------
# Load the gapminder 1997 data
gm97 = read_csv('data/gapminder_1997.csv')Rows: 142 Columns: 6
── Column specification ───────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
Delimiter: ","
chr (2): country, continent
dbl (4): year, pop, lifeExp, gdpPercap
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.Remember, with the cursor on this line we can click Run,
or we can type Ctrl+Enter. We should see some
output in the Console pane as well as gm97 in the
Environment pane. We’ll explore the resulting data in later lessons.
Checkpoint
Let’s break down this command:
gm97is the variable name we’re giving to the data we read in.=is the assignment operator which assigns the object on the right to the name on the left.read_csv()is a function intidyversethat reads CSV files.data/gapminder_1997.csvis the argument toread_csv()that specifies the file to read.
The output of read_csv() in the Console pane gives
information such as the dimensions of the data, the delimiter of the
file, and how the columns of the data were interpreted.
Note: The assignment operator
You may have seen another assignment operator,
<-, which is idiosyncratic to R. We leave as an exercise to the learner to look up the edge cases of when to use<-vs=. For now, we will use=as the assignment operator, which is more common in other programming languages.
Basic R data types
In the output of read_csv(), the country
and continent columns were intepreted as character strings
(chr) and the year, pop,
lifeExp, and gdpPercap columns were
interpreted as numbers (dbl). This begs the question of the
data types available in R. The basic data types in R are:
| Mode (abbreviation) | Type of data | Example |
|---|---|---|
| Numeric (num) | Decimals, integers, etc. | 1.0, 3.14, -2.5,
10, etc. |
| Character (chr) | Sequence of letters or numbers. | "Hi", 'Hi', "1",
etc. |
| Factor (fct) | Categorical values. | Months of the year. |
| Logical | Boolean values | TRUE, FALSE, T,
F, etc. |
Assigning values to objects
Throughout this workshop we’ll be assigning names to objects and manipulating them. The names we give to objects can either make our lives easier or harder. Let’s start by describing good practices, and then we’ll give some examples of bad practices.
The good
- Use brief, descriptive names.
- Separate words with underscores.
# -------------------------------------------------------------------------
# Examples of good variable names, and writing over an existing variable
age = 26
age[1] 26wizard_name = 'Tom Riddle'
wizard_name[1] "Tom Riddle"wizard_name = 'Harry Potter'
wizard_name[1] "Harry Potter"The bad
- No spaces.
- Don’t start with numbers.
- Don’t use proteted names, like
if,else,for, etc. (see here for complete list).
# -------------------------------------------------------------------------
# Error: Example of variable name with space
favorite number = 12Error in parse(text = input): <text>:4:10: unexpected symbol
3:
4: favorite number
^# -------------------------------------------------------------------------
# Error: Example of variable name beginning with number
1number = 3Error in parse(text = input): <text>:4:2: unexpected symbol
3:
4: 1number
^Be mindful
- Object names are case-sensitive. This means that
name,Name, andNAMEare three distinct objects. Imagine what confusion you could create! - Words in object names can also be separated by camelCase
(e.g.
objectName), but be consistent.
# -------------------------------------------------------------------------
# Example of case-sensitivity of variable names
Flower = 'marigold'
Flower[1] "marigold"flower = 'rose'
flower[1] "rose"# -------------------------------------------------------------------------
# Example of camelCase variable name
favoriteNumber = 12
favoriteNumber[1] 12Notice that with each assignment, the object appears in the
Environment pane. Also notice that by assigning wizard_name
twice, the value becomes the last assigned value, overwriting our
initial assignment. Also notice that if we evaluate the name of an
object, it is printed in the Console pane. We will use this
pattern repeatedly.
Checkpoint
Calling functions
Earlier we ran the code
gm97 = read_csv('data/gapminder_1997.csv'). As we said
before, read_csv() is a function and
'data/gapminder_1997.csv' is an argument to that function.
What happens if we just do:
# -------------------------------------------------------------------------
# Example of a function that needs arguments to function
read_csv()Error in read_csv(): argument "file" is missing, with no defaultWe get an error in the Console pane. The key part of the message is “argument ‘file’ is missing, with no default”. In other words, this function needs to be told what to read because there is no default.
Not every function needs arguments, but many do. Try the following functions:
# -------------------------------------------------------------------------
# Examples of functions with no required arguments
Sys.Date()[1] "2025-10-03"getwd()[1] "/home/workshop/rcavalca/workshop-intro-r-rstudio/source"# -------------------------------------------------------------------------
# Example of a function with multiple arguments
round(3.1415, 2)[1] 3.14Notice that we threw in round() which actually takes two
arguments. How could we have known that?
Getting help with functions
When a function is unfamiliar, we’ll often look at the manual page
for the function to understand what arguments are required, what it
does, and what it outputs. By prepending a ? in front of a
function name, you can access the manual page.
# -------------------------------------------------------------------------
# Put a "?" in front of a function to see it's manual page
?roundThe help page for round() tells us the function does
essentially what we’d expect, and gives some other related functions.
Note also that the arguments section gives us the names of the arguments
and what is expected of them. There is often a Details section to
describe nuances, and a Value section to describe the output. Finally,
there is an Examples section which gives examples of how to run the
code.
Arguments
When we called round(3.1415, 2) it seemed like the first
argument is the thing we want to round, and the second argument is how
many digits we want. That tracks when we look at ?round. R
can evaluate arguments of a function based on their
position, as we just saw. However, the preferred way to
call a function is to use the names of the arguments, as in:
# -------------------------------------------------------------------------
# Example of named arguments
round(x = 3.14159, digits = 2)[1] 3.14Calling a function, and using named arguments, increases the readability of the code and reduces the chance of error, especially with complex functions having many arguments.
Searching for functions
Prepending a ? in front a function name to find out more
about the function requires knowing the name of the function beforehand.
That won’t always be the case so there are a couple ways to search for R
functions.
- Search the internet for “R function that does X”.
- Use
help.search(), as inhelp.search('Chi-squared test')
Note that in the results of help.search() we see things
like, stats::chisq.test. Here the :: is R
notation for package_name::function.
Checkpoint
Errosr happn
We already assigned some variables that resulted in errors. There will be plenty of more of those to come; they’re a normal part of coding and they are an opportunity to learn! To that end, let’s make some mistakes together.
# -------------------------------------------------------------------------
# Example of not closing quotes
read_csv('data/gapminder_1997.csv)# -------------------------------------------------------------------------
# Example of not closing parentheses
round(3.1415, 2In both cases, the Console displays a + to indicate that
R is waiting for more input. To get out of this state, we can press
Esc, and try again.
Handling errors
The key to correcting errors is understanding what went wrong. Sometimes R can help, while other times it seems willfully obtuse.
- Immediately check the spelling of the command; most errors are caused by typos.
- Read the error message to try to understand what might have gone wrong.
- Check that the objects going into the function are what you expect.
If you’re still stuck as to why an error occurred (something we all encounter), reach out for help. For the workshop, please post the question in Slack with the following information:
- The exact command that caused the error.
- The exact error message that resulted.
This way we’ll more quickly be able to diagnose the problem.
Base R and the tidyverse
If you’ve used R before, you may have learned commands that are
different what we’ll learn in this workshop. We’ll focus on functions
from the tidyverse, a collection of R packages designed to
work well together and and offer many features that aren’t part of a
fresh install of R (that is, “base R”). Generally the
tidyverse helps us write code that is easy to read and
maintain, as we’ll see.
The tidyverse is geared for data in the form of tables,
and it is very good at manipulating, summarizing, and
visualizing such data. However, data occurs in a variety of other shapes
and forms. In particular, in a bioinformatics context, the Bioconductor
repository of packages utilize data types that are not tables, and
therefore do not always work well with tidyverse functions.
We’ll see clearer examples of this in the RNA-seq Demystified workshop,
and you will undoubtedly encounter many examples in the future.
Some people ask “Should I learn tidyverse or base R?” and we think
that rather than either/or, it’s better to think of both/and. Knowing
base R and its approach will help in some contexts, while knowing
tidyverse will help in others.
Cheatsheets!
The tidyverse packages have excellent cheatsheets that
describe the functionality and usage of the packages. You can find them
in RStudio by going to the “Help” menu and selecting “Cheat Sheets”. The
two that will be most helpful in this workshop are “Data Visualization
with ggplot2” and “Data Transformation with dplyr”.
Objectives
- Introduce R and RStudio.
- Learn how to read in a csv file.
- Discuss object naming conventions and types.
- Discuss functions and parameters.
- Learn how to get help with functions.
- Get comfortable with errors and asking for help.
| Previous lesson | Top of this lesson | Next lesson |
|---|