Reproducible Computing Workshop
UM Bioinformatics Core
Wrapping up
We hope you now have more familiarity with key patterns, approaches, and tools that can enable more computational reproducibility in your research. Please remember that reproducibility exists on a continuum:
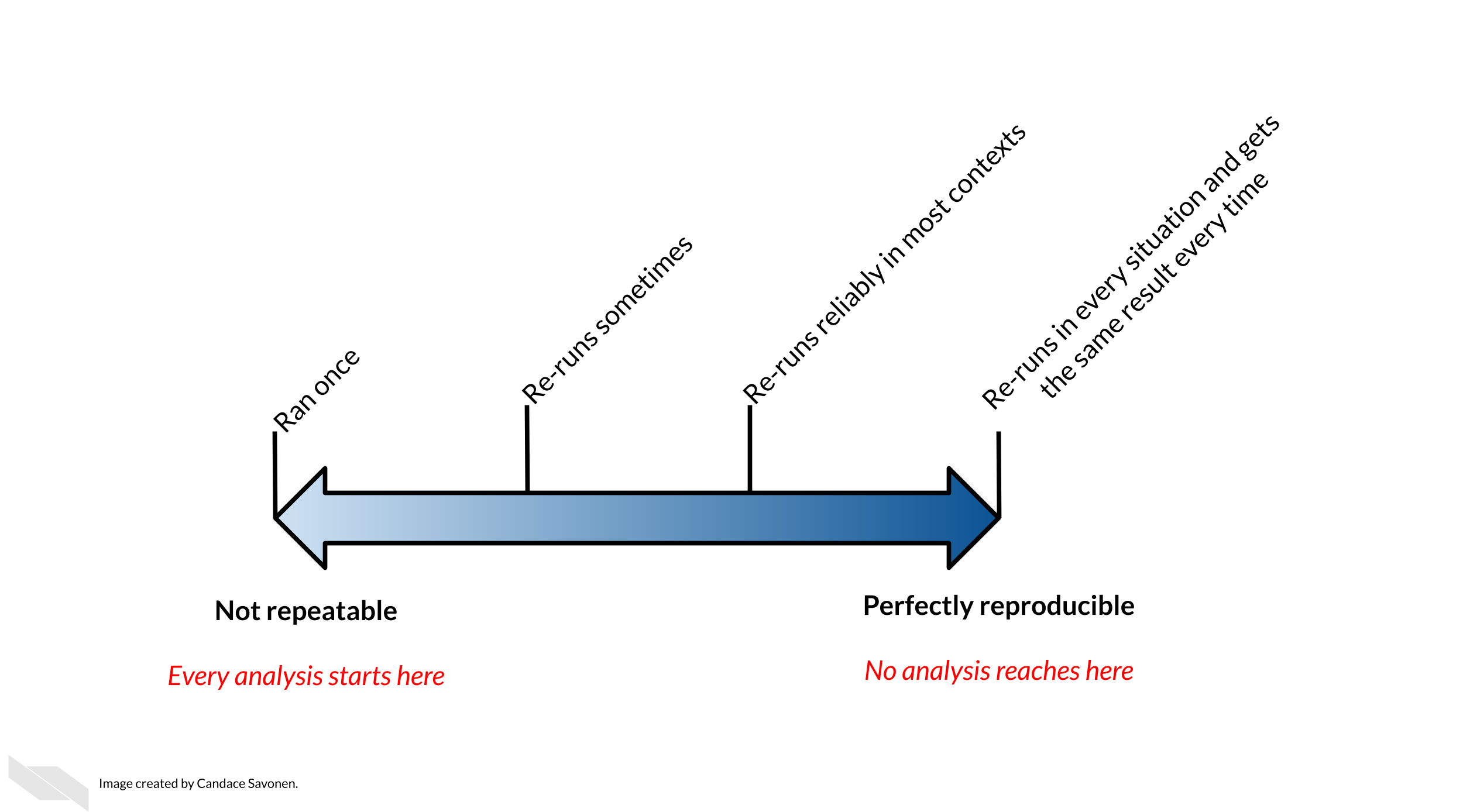 Sourced from Advanced Reproducibility in Cancer Informatics / The
Johns Hopkins Data Science Lab —
Sourced from Advanced Reproducibility in Cancer Informatics / The
Johns Hopkins Data Science Lab —
Housekeeping
Please take our optional post-workshop survey. (5-10 minutes)
We will email you a link to the final session recordings by end of next week.
The website/notes for this workshop will be available.
The UM Bioinformatics Core Workshop Slack channel will be available for 90 days.
Looking ahead
Workshop environment
This Great Lakes account will will be available to you until 6/30/24. Following that, you will be removed this account and any content inside the workshop Turbo or DataDen will be removed.
You can download files from your workshop home dir using your terminal/command line window as below. (You will need to substitute your actual uniqname and type your password when prompted.)
mkdir computational-reproducibility-workshop cd computational-reproducibility-workshop scp -r greatlakes-xfer.arc-ts.umich.edu:/nfs/turbo/umms-bioinf-wkshp/workshop/home/YOUR_USERNAME .Please keep in mind that this workshop environment is optimized for the exercises in this particular workshop but is likely unsuitable for analyzing your own datasets. In particular:
It is not sized for compute intensive operations or large storage.
It is not secured for sensitive data of any kind.
University of Michigan Resources
Continued learning and support
Making research reproducibile is a learning process. Resources to consider:
- UM CoderSpaces “office hours” and UM CoderSpaces Slack workspace. (See “Useful Resources” section in above page for instructions on how to join and access the CoderSpaces Slack workspace.)
- Upcoming UM Advanced Research Computing workshops.
- Videos on getting started with Great Lakes.
- For more intro lessons and workshops in Bash / Git / R / Python : Software Carpentry and the UM Software Carpentry Group.
- Advanced Reproducibility in Cancer Informatics from The Johns Hopkins Data Science Lab
Thank you

Thank you to/from the workshop team
 |
 |
 |
|
|---|---|---|---|
| Chris | Marci | Travis | |
 |
 |
||
| Dana | Raymond |
Thank you for participating in our workshop. We welcome your questions and feedback now and in the future.
Bioinformatics Workshop Team
bioinformatics-workshops@umich.edu
https://medresearch.umich.edu/office-research/about-office-research/biomedical-research-core-facilities/bioinformatics-core