Exercises - Intro to Single-Cell Analysis
UM Bioinformatics Core Workshop Team
Workflow Overview
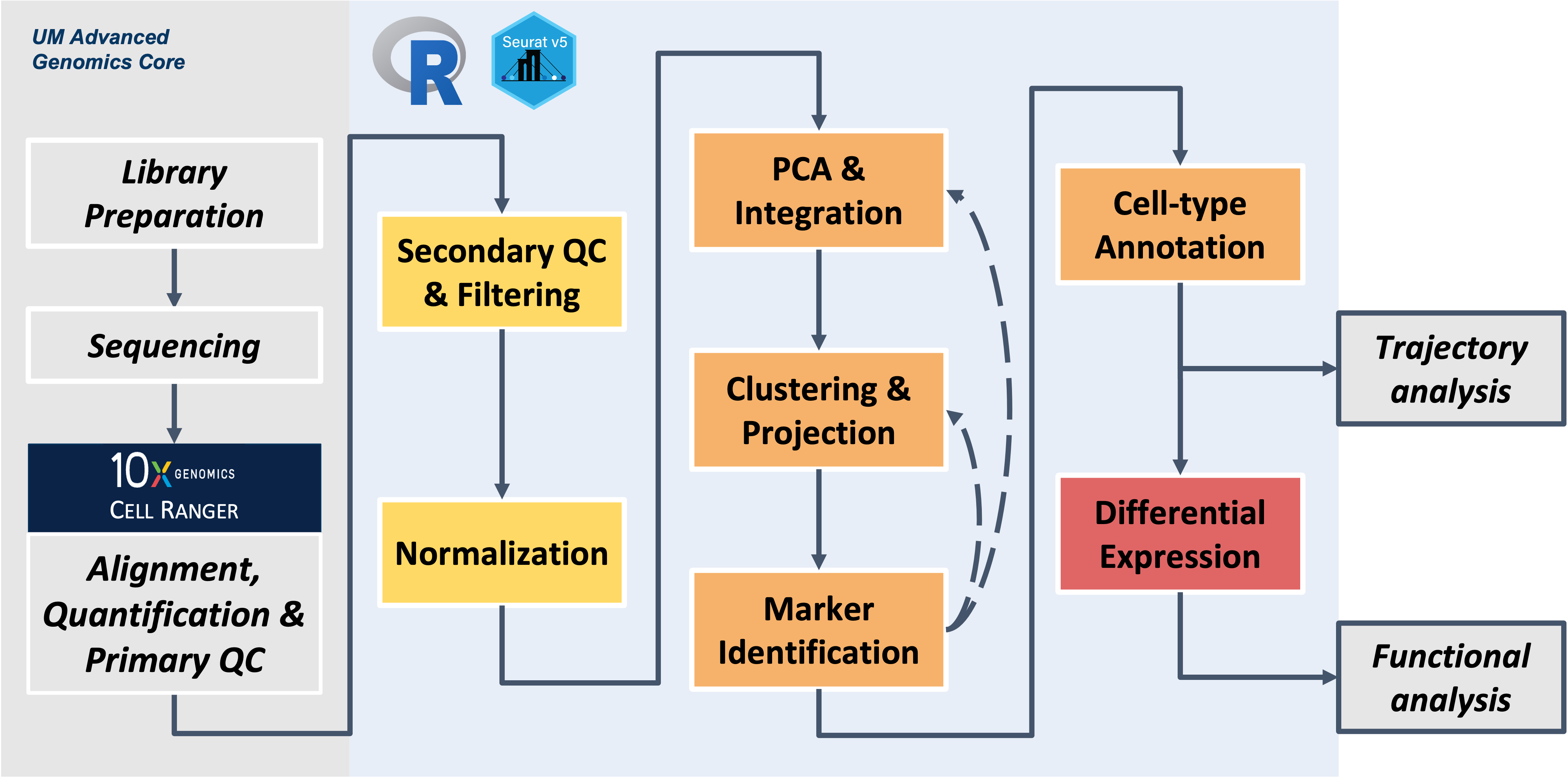
Workshop exercises
Build on the content and analysis steps covered in the workshop
sessions by working through these independent exercises. Note -
if you work on the exercises make sure to restart R session to clear out
environment before closing the window (like we have at the end of each
session) to avoid lags when logging in the next day
Reminder: RStudio, Seurat, and Memory Management
R and RStudio are designed to grab computer memory (RAM) when necessary and release that memory when it’s no longer needed. R does this with a fancy computer science technique called garbage collection.
The R garbage collector is very good for small objects and simple analyses, but complex single-cell analysis can overwhelm it. So don’t rely on R/RStudio’s built in garbage collection and session management for single-cell analysis. Instead, do the following:
- Save intermediate versions of the Seurat object as you go along
using
saveRDS(). This gives you “savepoints” so if you need to backtrack, you don’t have to go back to the beginning. - Save ggplots into image files as you go along. Periodically clear
ggplot objects from the R environment.
- At the end of each day, wherever you are in the analysis, explicitly
save the Seurat object and “power down” your session.
- The next day, you can load the Seruat object using readRDS().
This simple pattern will make your code more reproducible and your RStudio session more reliable.
Day 1
You don’t need to have a fresh environment for these exercises. All variable names will be different from the lessons. Load the libraries (it’s okay if they’re already loaded) and the unfiltered Seurat object with the following code:
# =========================================================================
# Independent Exercise - Day 1 Startup
# =========================================================================
# After restarting our R session, load the required libraries & input data
library(Seurat)
library(BPCells)
library(tidyverse)
# Use provided copy of integrated data
exso = readRDS('./inputs/prepared_data/rdata/geo_so_unfiltered.rds')
# Add percent.mt column to meta.data
exso$percent.mt = PercentageFeatureSet(exso, pattern = '^mt-')
## NOTE - BEFORE STOPPING WORK ON THE EXERCISES REMEMBER TO POWER DOWN AND RESTART R SESSION !!!!Let’s practice filtering cells based on different criteria than we’re using in the lessons.
Day 1 Exercise 1
Subset exso so that all cells have
nFeature_RNA greater than or equal to 1000. How many cells
remain?
Hint 1: For a reminder of how to subset a Seurat object, see the “Removing low-quality cells” section of the Secondary QC and Filtering module. Hint 2: For a reminder, of how to count the number cells per sample, see the “Cell counts” section of the Secondary QC and Filtering module.
Day 1 Exercise 2
Subset exso so that all cells have
nCount_RNA greater than or equal to 5000. How many cells
remain?
Day 1 Exercise 3
Subset exso so that all cells have
percent.mt less than 10%. How many cells remain?
Day 1 Exercise 4
Subset exso so that each of the previous three
conditions are satisfied. How many cells remain?
Day 1 solutions
Scripts
# =========================================================================
# Independent Exercise - Day 1 Startup
# =========================================================================
# After restarting our R session, load the required libraries & input data
library(Seurat)
library(BPCells)
library(tidyverse)
# Load the unfiltered version and give it a new variable name
exso = readRDS('./inputs/prepared_data/rdata/geo_so_unfiltered.rds')
# Add percent.mt column to meta.data
exso$percent.mt = PercentageFeatureSet(exso, pattern = '^mt-')
## NOTE - BEFORE STOPPING WORK ON THE EXERCISES REMEMBER TO POWER DOWN AND RESTART R SESSION !!!!
# Exercise 1
exso = subset(exso, nFeature_RNA >= 1000)
ex1 = exso@meta.data %>% count(orig.ident, name = 'postfilter_cells')
ex1
# Exercise 2
exso = subset(exso, nCount_RNA >= 1000)
ex2 = exso@meta.data %>% count(orig.ident, name = 'postfilter_cells')
ex2
# Exercise 3
exso = subset(exso, percent.mt < 10)
ex3 = exso@meta.data %>% count(orig.ident, name = 'postfilter_cells')
ex3
# Exercise 4
exso = subset(exso, nFeature_RNA >= 1000 & nCount_RNA >= 1000 & percent.mt < 10)
ex4 = exso@meta.data %>% count(orig.ident, name = 'postfilter_cells')
ex4Full output
# =========================================================================
# Independent Exercise - Day 1 Startup
# =========================================================================
# After restarting our R session, load the required libraries & input data
library(Seurat)
library(BPCells)
library(tidyverse)
# Load the unfiltered version and give it a new variable name
exso = readRDS('./inputs/prepared_data/rdata/geo_so_unfiltered.rds')
# Add percent.mt column to meta.data
exso$percent.mt = PercentageFeatureSet(exso, pattern = '^mt-')
## NOTE - BEFORE STOPPING WORK ON THE EXERCISES REMEMBER TO POWER DOWN AND RESTART R SESSION !!!!
# Exercise 1
exso = subset(exso, nFeature_RNA >= 1000)
ex1 = exso@meta.data %>% count(orig.ident, name = 'postfilter_cells')
ex1 orig.ident postfilter_cells
1 HODay0replicate1 937
2 HODay0replicate2 545
3 HODay0replicate3 992
4 HODay0replicate4 833
5 HODay21replicate1 1854
6 HODay21replicate2 1094
7 HODay21replicate3 1093
8 HODay21replicate4 2288
9 HODay7replicate1 3756
10 HODay7replicate2 4212
11 HODay7replicate3 4207
12 HODay7replicate4 3445# Exercise 2
exso = subset(exso, nCount_RNA >= 1000)
ex2 = exso@meta.data %>% count(orig.ident, name = 'postfilter_cells')
ex2 orig.ident postfilter_cells
1 HODay0replicate1 937
2 HODay0replicate2 545
3 HODay0replicate3 992
4 HODay0replicate4 833
5 HODay21replicate1 1854
6 HODay21replicate2 1094
7 HODay21replicate3 1093
8 HODay21replicate4 2288
9 HODay7replicate1 3756
10 HODay7replicate2 4212
11 HODay7replicate3 4207
12 HODay7replicate4 3445# Exercise 3
exso = subset(exso, percent.mt < 10)
ex3 = exso@meta.data %>% count(orig.ident, name = 'postfilter_cells')
ex3 orig.ident postfilter_cells
1 HODay0replicate1 886
2 HODay0replicate2 535
3 HODay0replicate3 976
4 HODay0replicate4 821
5 HODay21replicate1 1793
6 HODay21replicate2 1059
7 HODay21replicate3 1052
8 HODay21replicate4 2179
9 HODay7replicate1 3574
10 HODay7replicate2 4057
11 HODay7replicate3 4069
12 HODay7replicate4 3299# Exercise 4
exso = subset(exso, nFeature_RNA >= 1000 & nCount_RNA >= 1000 & percent.mt < 10)
ex4 = exso@meta.data %>% count(orig.ident, name = 'postfilter_cells')
ex4 orig.ident postfilter_cells
1 HODay0replicate1 886
2 HODay0replicate2 535
3 HODay0replicate3 976
4 HODay0replicate4 821
5 HODay21replicate1 1793
6 HODay21replicate2 1059
7 HODay21replicate3 1052
8 HODay21replicate4 2179
9 HODay7replicate1 3574
10 HODay7replicate2 4057
11 HODay7replicate3 4069
12 HODay7replicate4 3299Day 2
Since we are working with larger object sizes, it’s best to start with a fresh session. All variable names will be different from the lessons. Load the libraries (it’s okay if they’re already loaded) and the integrated Seurat object with the following code:
# =========================================================================
# Independent Exercise - Day 2 Startup
# =========================================================================
# After restarting our R session, load the required libraries & input data
library(Seurat)
library(BPCells)
library(tidyverse)
# Use provided copy of integrated data
exso2 = readRDS('inputs/prepared_data/rdata/geo_so_sct_integrated.rds')
exso2 # check that object loaded
## NOTE - BEFORE STOPPING WORK ON THE EXERCISES REMEMBER TO POWER DOWN AND RESTART R SESSION !!!!Day 2 Exercise 1 - Clustering with reduced number of PCs
Test how the clustering results would change if you used fewer or more PCs when clustering these data.
# -------------------------------------------------------------------------
# Testing fewer PCs for clustering
# look at elbow plot to check PCs and consider alternatives to the number selected for main materials
ElbowPlot(exso2, ndims = 50, reduction = 'unintegrated.sct.pca')
# select alternative value to try (choose a number <10 or >10 PCs)
pcs = # Your value here #
# generate nearest neighbor (graph), using selected number of PCs
exso2 = FindNeighbors(exso2, dims = 1:pcs, reduction = 'integrated.sct.rpca')After generating clustering, set resolution parameter - remember this only impacts the how the boundaries of the neighbors are drawn, not the underlying NN graph/structure.
# -------------------------------------------------------------------------
# Testing resolution options to see impact
# start with one resolution
res = # Your value here #
# generate clusters, using `pcs` and `res` to make a custom cluster name that will be added to the metadata
exso2 = FindClusters(exso2, resolution = res,
cluster.name = paste0('int.sct.rpca.clusters', res))
# look at meta.data to see cluster labels
head(exso2@meta.data)
# run UMAP reduction to prepare for plotting
exso2 = RunUMAP(exso2, dims = 1:pcs,
reduction = 'integrated.sct.rpca',
reduction.name = paste0('umap.integrated.sct.rpca_alt', res))
# check object to see if named reduction was added
exso2Challenge 1: How could we generate clusters for
multiple alternative resolutions (e.g. 0.4, 0.8, & 1.0)?
Hint - for loops
can be useful for iteration in many programming languages, including
R.
Day 2 Exercise 2 - Plotting alternative clustering results
After generating our clustering results it’s time to visualize them - create a UMAP plot, ensuring that you generate a UMAP reduction for the appropriate reduction
Hint 1 - Remember the previous code chunk used
paste0('umap.integrated.sct.rpca_alt', res)) for the
cluster name parameter.
Hint 2 - For a reminder of how to generate a UMAP plot, look at the Visualizing and evaluating clustering from the Clustering and Projection module.
Challenge 2: Create UMAP plots for the alternative resolutions (e.g. 0.4, 0.8, & 1.0) generated in the previous challenge.
Check-in Questions - UMAP for alternative PCs
How does the UMAP plot look when fewer or more PCs are used? Do you notice any relationship between the PC parameter and resolution parameter?
Clean up session
Before closing out the window, make sure to clear environment and restart R session to manage the memory usage
## ----------------------------------------------------------
## Clean up session, including any plot objects
rm(list=names(which(unlist(eapply(.GlobalEnv, is.ggplot)))));
gc()
## Save copy of Seurat object in current state to file
saveRDS(exso2, file = paste0('results/rdata/geo_so_sct_integrated_exercise.rds'))
rm(exso2)
gc()
Day 2 solutions
Scripts
# =========================================================================
# Independent Exercise - Day 2 Startup
# =========================================================================
# After restarting our R session, load the required libraries & input data
library(Seurat)
library(BPCells)
library(tidyverse)
# Use provided copy of integrated data
exso2 = readRDS('inputs/prepared_data/rdata/geo_so_sct_integrated.rds')
exso2 # check that object loaded
### Day 2 Exercise 1 - Clustering with reduced number of PCs
# -------------------------------------------------------------------------
# Testing fewer PCs for clustering
# look at elbow plot to check PCs and consider alternatives to the number selected for main materials
ElbowPlot(exso2, ndims = 50, reduction = 'unintegrated.sct.pca')
# select alternative value to try (can choose a number <10 or >10 PCs)
pcs = 6
# generate nearest neighbor (graph), using selected number of PCs
exso2 = FindNeighbors(exso2, dims = 1:pcs, reduction = 'integrated.sct.rpca')
# -------------------------------------------------------------------------
# start with one resolution
res = 0.2
# generate clusters, using `pcs` and `res` to make a custom cluster name that will be added to the metadata
exso2 = FindClusters(exso2, resolution = res,
cluster.name = paste0('int.sct.rpca.clusters', res))
# look at meta.data to see cluster labels
head(exso2@meta.data)
# run UMAP reduction to prepare for plotting
exso2 = RunUMAP(exso2, dims = 1:pcs,
reduction = 'integrated.sct.rpca',
reduction.name = paste0('umap.integrated.sct.rpca_alt', res))
# check object to see if named reduction was added
exso2
## Challenge 1 - solution option
# -------------------------------------------------------------------------
# use for loop to generate clustering with alternative resolutions
for(i in c(0.4, 0.8, 1.0)){
exso2 = FindClusters(exso2, resolution = i,
cluster.name = paste0('int.sct.rpca.clusters', i))
# look at meta.data to see cluster labels
head(exso2@meta.data)
# run UMAP reduction to prepare for plotting
exso2 = RunUMAP(exso2, dims = 1:pcs,
reduction = 'integrated.sct.rpca',
reduction.name = paste0('umap.integrated.sct.rpca_alt', i))
# check object to see if multiple cluster resolutions are added
head(exso2@meta.data)
}
### Day 2 Exercise 2 - Plotting alternative clustering results
# -------------------------------------------------------------------------
# plot clustering results
post_integration_umap_clusters_testing =
DimPlot(exso2, group.by = paste0('int.sct.rpca.clusters', res), label = TRUE,
reduction = paste0('umap.integrated.sct.rpca_alt', res)) + NoLegend()
post_integration_umap_clusters_testing # look at plot
# output to file, including the number of PCs and resolution used to generate results
ggsave(filename = paste0('results/figures/umap_int_sct_clusters_exercise_',
pcs,'PC.',res,'res','.png'),
plot = post_integration_umap_clusters_testing,
width = 8, height = 6, units = 'in')
## Challenge 2 - solution option
# -------------------------------------------------------------------------
# use for loop to visualize clustering across tested resolutions
post_integration_umap_plots <- c()
for(i in c(0.4, 0.8, 1.0)){
res_type = paste0("res_", i)
post_integration_umap_plots[[res_type]] =
DimPlot(exso2, group.by = paste0('int.sct.rpca.clusters', i), label = TRUE,
reduction = paste0('umap.integrated.sct.rpca_alt', i)) + NoLegend()
}
# look at plots for each resolution stored in list
post_integration_umap_plots
# remove plot list to clean up session
rm(post_integration_umap_plots)
## ----------------------------------------------------------
## Clean up session, including any plot objects
rm(list=names(which(unlist(eapply(.GlobalEnv, is.ggplot)))));
gc()
## Save copy of Seurat object in current state to file
saveRDS(exso2, file = paste0('results/rdata/geo_so_sct_clustered_exercise.rds'))
rm(exso2)
gc()Full output
# =========================================================================
# Independent Exercise - Day 2 Startup
# =========================================================================
# After restarting our R session, load the required libraries & input data
library(Seurat)
library(BPCells)
library(tidyverse)
# Use provided copy of integrated data
exso2 = readRDS('inputs/prepared_data/rdata/geo_so_sct_integrated.rds')
exso2 # check that object loadedAn object of class Seurat
46957 features across 31559 samples within 2 assays
Active assay: SCT (20468 features, 3000 variable features)
3 layers present: counts, data, scale.data
1 other assay present: RNA
2 dimensional reductions calculated: unintegrated.sct.pca, integrated.sct.rpca### Day 2 Exercise 1 - Clustering with reduced number of PCs
# -------------------------------------------------------------------------
# Testing fewer PCs for clustering
# look at elbow plot to check PCs and consider alternatives to the number selected for main materials
ElbowPlot(exso2, ndims = 50, reduction = 'unintegrated.sct.pca')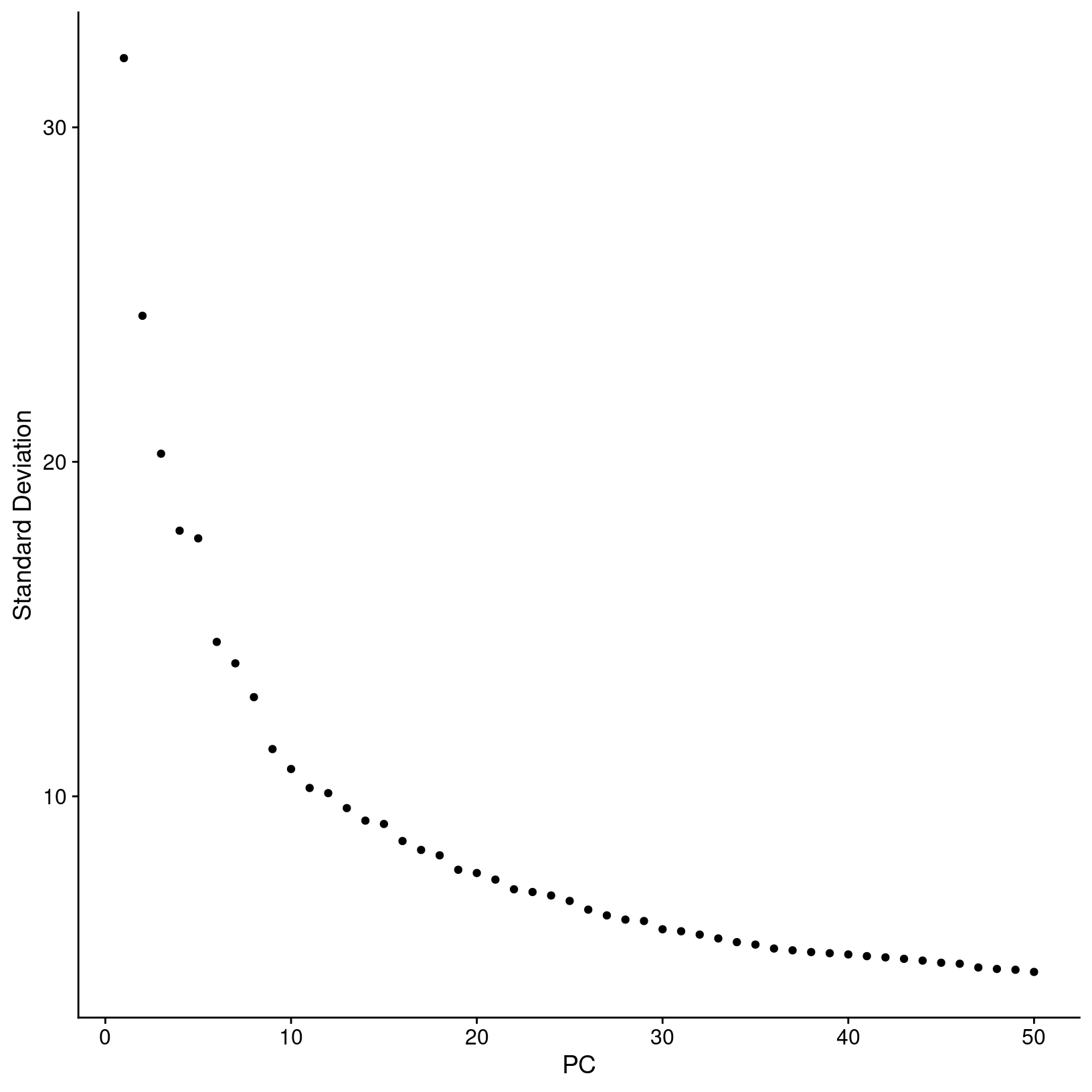
# select alternative value to try (can choose a number <10 or >10 PCs)
pcs = 6
# generate nearest neighbor (graph), using selected number of PCs
exso2 = FindNeighbors(exso2, dims = 1:pcs, reduction = 'integrated.sct.rpca')Computing nearest neighbor graphComputing SNN# -------------------------------------------------------------------------
# start with one resolution
res = 0.2
# generate clusters, using `pcs` and `res` to make a custom cluster name that will be added to the metadata
exso2 = FindClusters(exso2, resolution = res,
cluster.name = paste0('int.sct.rpca.clusters', res))Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 31559
Number of edges: 950342
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.9612
Number of communities: 12
Elapsed time: 3 seconds# look at meta.data to see cluster labels
head(exso2@meta.data) orig.ident nCount_RNA nFeature_RNA condition time replicate percent.mt nCount_SCT nFeature_SCT int.sct.rpca.clusters0.2 seurat_clusters
HODay0replicate1_AAACCTGAGAGAACAG-1 HODay0replicate1 10234 3226 HO Day0 replicate1 1.240962 6062 2867 0 0
HODay0replicate1_AAACCTGGTCATGCAT-1 HODay0replicate1 3158 1499 HO Day0 replicate1 7.536415 4607 1509 0 0
HODay0replicate1_AAACCTGTCAGAGCTT-1 HODay0replicate1 13464 4102 HO Day0 replicate1 3.112002 5314 2370 0 0
HODay0replicate1_AAACGGGAGAGACTTA-1 HODay0replicate1 577 346 HO Day0 replicate1 1.559792 3877 1031 7 7
HODay0replicate1_AAACGGGAGGCCCGTT-1 HODay0replicate1 1189 629 HO Day0 replicate1 3.700589 4166 915 0 0
HODay0replicate1_AAACGGGCAACTGGCC-1 HODay0replicate1 7726 2602 HO Day0 replicate1 2.938131 5865 2588 0 0# run UMAP reduction to prepare for plotting
exso2 = RunUMAP(exso2, dims = 1:pcs,
reduction = 'integrated.sct.rpca',
reduction.name = paste0('umap.integrated.sct.rpca_alt', res))Warning: The default method for RunUMAP has changed from calling Python UMAP via reticulate to the R-native UWOT using the cosine metric
To use Python UMAP via reticulate, set umap.method to 'umap-learn' and metric to 'correlation'
This message will be shown once per session21:30:02 UMAP embedding parameters a = 0.9922 b = 1.11221:30:02 Read 31559 rows and found 6 numeric columns21:30:02 Using Annoy for neighbor search, n_neighbors = 3021:30:02 Building Annoy index with metric = cosine, n_trees = 500% 10 20 30 40 50 60 70 80 90 100%[----|----|----|----|----|----|----|----|----|----|**************************************************|
21:30:06 Writing NN index file to temp file /tmp/Rtmp0Y5yMq/file124dae097d3
21:30:06 Searching Annoy index using 1 thread, search_k = 3000
21:30:19 Annoy recall = 100%
21:30:20 Commencing smooth kNN distance calibration using 1 thread with target n_neighbors = 30
21:30:22 Initializing from normalized Laplacian + noise (using RSpectra)
21:30:25 Commencing optimization for 200 epochs, with 1214176 positive edges
21:30:25 Using rng type: pcg
21:30:38 Optimization finished# check object to see if named reduction was added
exso2An object of class Seurat
46957 features across 31559 samples within 2 assays
Active assay: SCT (20468 features, 3000 variable features)
3 layers present: counts, data, scale.data
1 other assay present: RNA
3 dimensional reductions calculated: unintegrated.sct.pca, integrated.sct.rpca, umap.integrated.sct.rpca_alt0.2## Challenge 1 - solution option
# -------------------------------------------------------------------------
# use for loop to generate clustering with alternative resolutions
for(i in c(0.4, 0.8, 1.0)){
exso2 = FindClusters(exso2, resolution = i,
cluster.name = paste0('int.sct.rpca.clusters', i))
# look at meta.data to see cluster labels
head(exso2@meta.data)
# run UMAP reduction to prepare for plotting
exso2 = RunUMAP(exso2, dims = 1:pcs,
reduction = 'integrated.sct.rpca',
reduction.name = paste0('umap.integrated.sct.rpca_alt', i))
# check object to see if multiple cluster resolutions are added
head(exso2@meta.data)
}Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 31559
Number of edges: 950342
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.9428
Number of communities: 20
Elapsed time: 3 seconds21:30:42 UMAP embedding parameters a = 0.9922 b = 1.112
21:30:42 Read 31559 rows and found 6 numeric columns
21:30:42 Using Annoy for neighbor search, n_neighbors = 30
21:30:42 Building Annoy index with metric = cosine, n_trees = 50
0% 10 20 30 40 50 60 70 80 90 100%
[----|----|----|----|----|----|----|----|----|----|
**************************************************|
21:30:46 Writing NN index file to temp file /tmp/Rtmp0Y5yMq/file124da1abc6065
21:30:46 Searching Annoy index using 1 thread, search_k = 3000
21:30:59 Annoy recall = 100%
21:31:00 Commencing smooth kNN distance calibration using 1 thread with target n_neighbors = 30
21:31:02 Initializing from normalized Laplacian + noise (using RSpectra)
21:31:05 Commencing optimization for 200 epochs, with 1214176 positive edges
21:31:05 Using rng type: pcg
21:31:18 Optimization finishedModularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 31559
Number of edges: 950342
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.9216
Number of communities: 28
Elapsed time: 3 seconds21:31:22 UMAP embedding parameters a = 0.9922 b = 1.112
21:31:22 Read 31559 rows and found 6 numeric columns
21:31:22 Using Annoy for neighbor search, n_neighbors = 30
21:31:22 Building Annoy index with metric = cosine, n_trees = 50
0% 10 20 30 40 50 60 70 80 90 100%
[----|----|----|----|----|----|----|----|----|----|
**************************************************|
21:31:26 Writing NN index file to temp file /tmp/Rtmp0Y5yMq/file124da9655b50
21:31:26 Searching Annoy index using 1 thread, search_k = 3000
21:31:39 Annoy recall = 100%
21:31:39 Commencing smooth kNN distance calibration using 1 thread with target n_neighbors = 30
21:31:41 Initializing from normalized Laplacian + noise (using RSpectra)
21:31:44 Commencing optimization for 200 epochs, with 1214176 positive edges
21:31:44 Using rng type: pcg
21:31:58 Optimization finishedModularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 31559
Number of edges: 950342
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.9136
Number of communities: 30
Elapsed time: 3 seconds21:32:02 UMAP embedding parameters a = 0.9922 b = 1.112
21:32:02 Read 31559 rows and found 6 numeric columns
21:32:02 Using Annoy for neighbor search, n_neighbors = 30
21:32:02 Building Annoy index with metric = cosine, n_trees = 50
0% 10 20 30 40 50 60 70 80 90 100%
[----|----|----|----|----|----|----|----|----|----|
**************************************************|
21:32:06 Writing NN index file to temp file /tmp/Rtmp0Y5yMq/file124da3bc483b2
21:32:06 Searching Annoy index using 1 thread, search_k = 3000
21:32:19 Annoy recall = 100%
21:32:19 Commencing smooth kNN distance calibration using 1 thread with target n_neighbors = 30
21:32:21 Initializing from normalized Laplacian + noise (using RSpectra)
21:32:24 Commencing optimization for 200 epochs, with 1214176 positive edges
21:32:24 Using rng type: pcg
21:32:37 Optimization finished### Day 2 Exercise 2 - Plotting alternative clustering results
# -------------------------------------------------------------------------
# plot clustering results
post_integration_umap_clusters_testing =
DimPlot(exso2, group.by = paste0('int.sct.rpca.clusters', res), label = TRUE,
reduction = paste0('umap.integrated.sct.rpca_alt', res)) + NoLegend()
post_integration_umap_clusters_testing # look at plot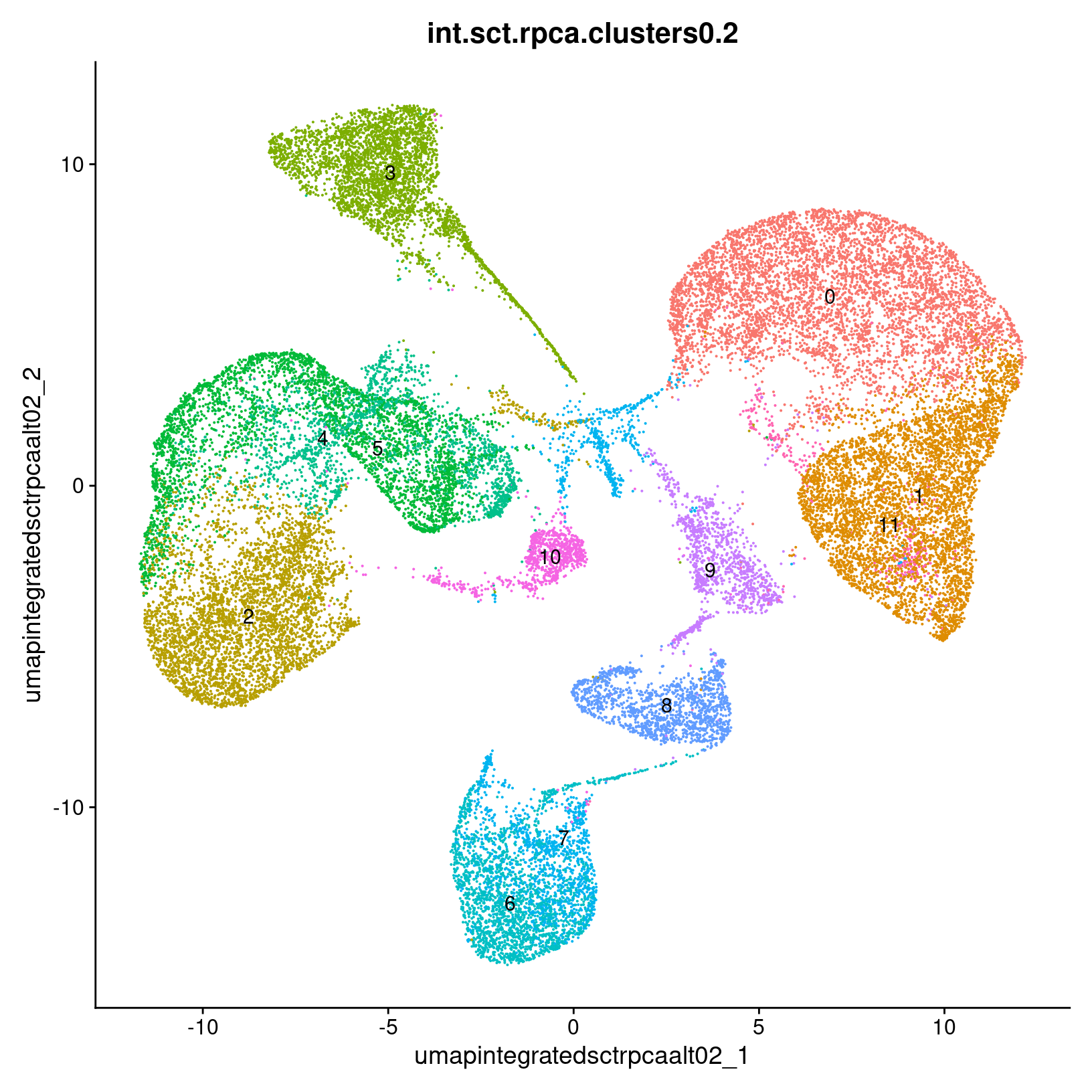
# output to file, including the number of PCs and resolution used to generate results
ggsave(filename = paste0('results/figures/umap_int_sct_clusters_exercise_',
pcs,'PC.',res,'res','.png'),
plot = post_integration_umap_clusters_testing,
width = 8, height = 6, units = 'in')
## Challenge 2 - solution option
# -------------------------------------------------------------------------
# use for loop to visualize clustering across tested resolutions
post_integration_umap_plots <- c()
for(i in c(0.4, 0.8, 1.0)){
res_type = paste0("res_", i)
post_integration_umap_plots[[res_type]] =
DimPlot(exso2, group.by = paste0('int.sct.rpca.clusters', i), label = TRUE,
reduction = paste0('umap.integrated.sct.rpca_alt', i)) + NoLegend()
}
# look at plots for each resolution stored in list
post_integration_umap_plots$res_0.4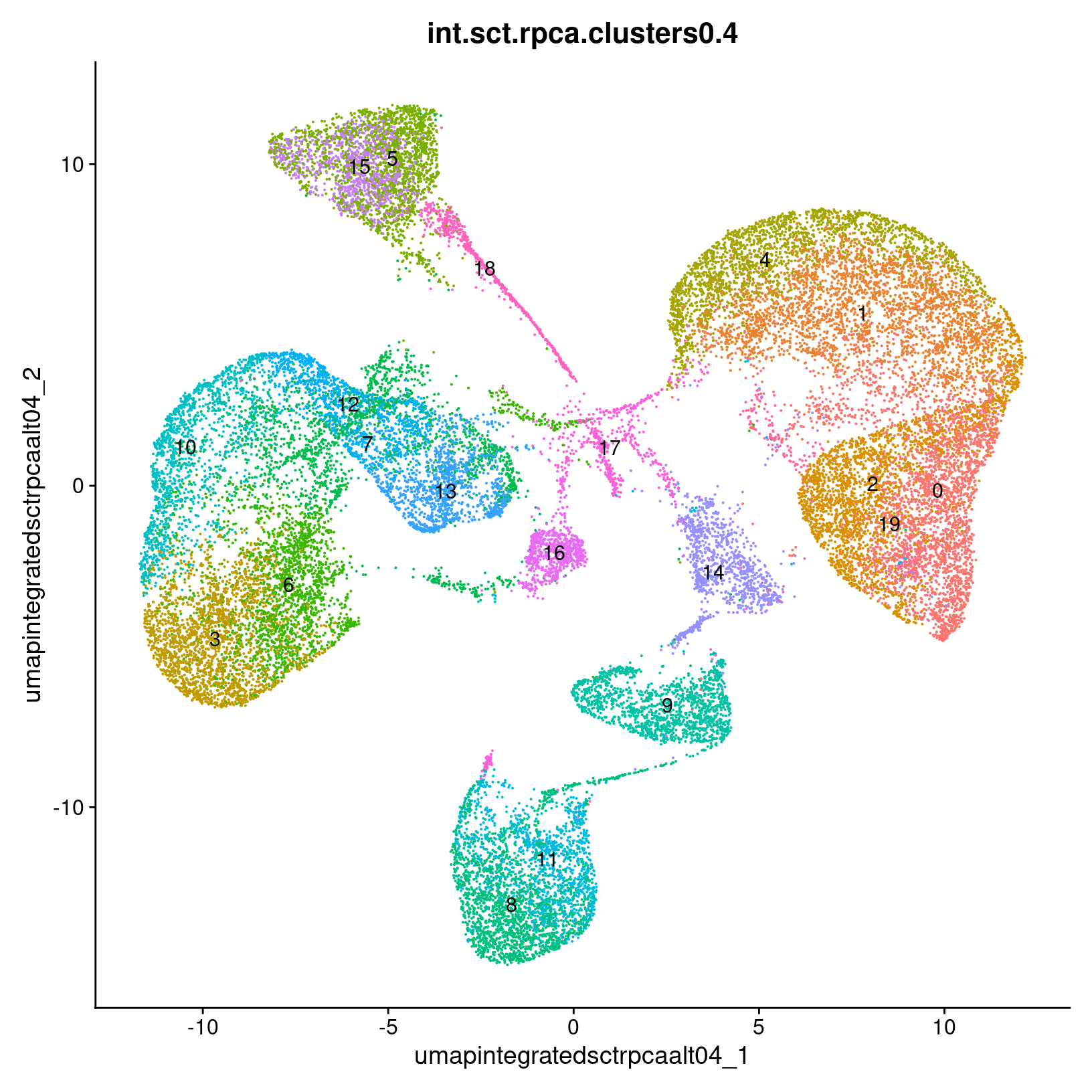
$res_0.8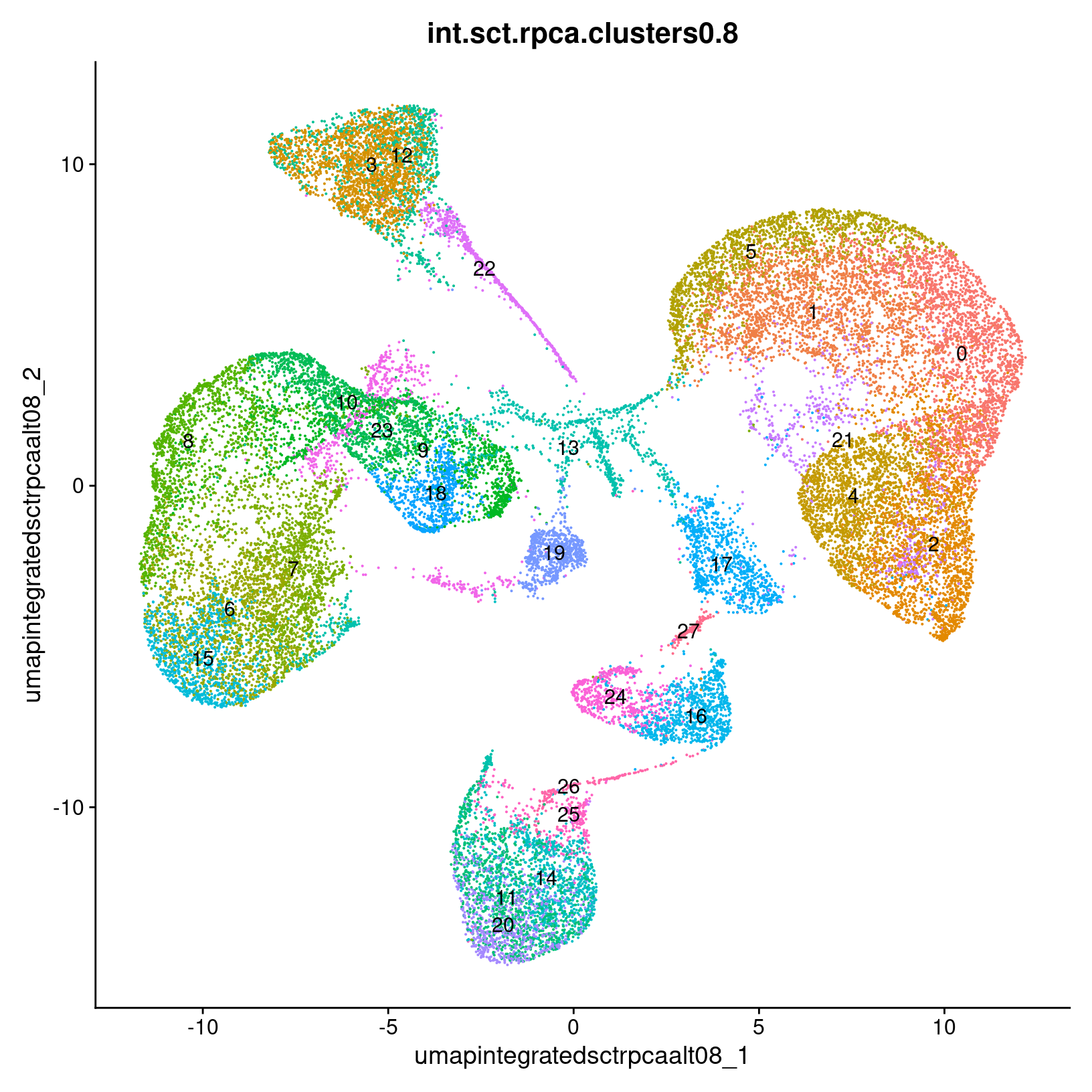
$res_1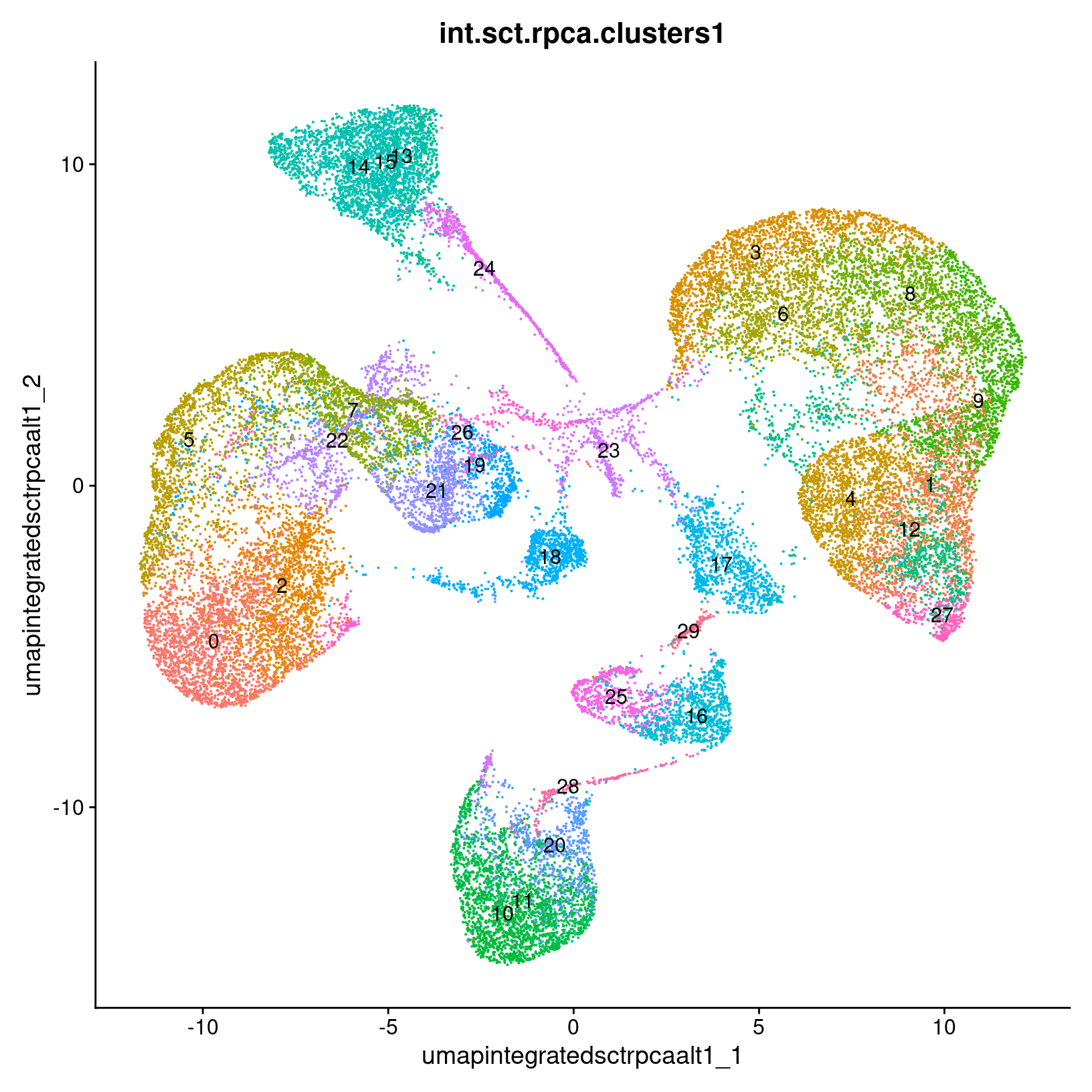
# remove plot list to clean up session
rm(post_integration_umap_plots)
## ----------------------------------------------------------
## Clean up session, including any plot objects
rm(list=names(which(unlist(eapply(.GlobalEnv, is.ggplot)))));
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 6967048 372.1 10752860 574.3 10752860 574.3
Vcells 384220421 2931.4 1067030484 8140.8 1333787743 10176.0## Save copy of Seurat object in current state to file
saveRDS(exso2, file = paste0('results/rdata/geo_so_sct_clustered_exercise.rds'))
rm(exso2)
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 6967076 372.1 10752860 574.3 10752860 574.3
Vcells 384236240 2931.5 1067030484 8140.8 1333787743 10176.0Day 3
Since we are working with larger object sizes, it’s best to start with a fresh session. All variable names will be different from the lessons. Load the libraries (it’s okay if they’re already loaded) and a Seurat object with alternative clustering using the following code:
# =========================================================================
# Independent Exercise - Day 3 Startup
# =========================================================================
# After restarting our R session, load the required libraries
library(Seurat)
library(BPCells)
library(tidyverse)
library(scCATCH)
# Load in seurat object with alternative clustering results from yesterday's exercises
exso3 = readRDS('results/rdata/geo_so_sct_integrated_exercise.rds')
exso3 # check that object loaded
## NOTE - BEFORE STOPPING WORK ON THE EXERCISES REMEMBER TO POWER DOWN AND RESTART R SESSION !!!!Before testing what marker genes and cell-type predictions are generated for an alternative clustering, start by ensuring the Seurat object is set up and has an expected identity set.
## ----------------------------------------------------------
# Check what identities are set
Idents(exso3) %>% head()
pcs = ## use same values as yesterday
res = ## use same values as yesterday
## Set identities to clustering for selected resolution
exso3 = SetIdent(exso3, value = paste0('int.sct.rpca.clusters', res))
Idents(exso3) %>% head()Day 3 Exercise 1 - examine marker genes for alternative clusters
Using the approach from the marker
identification module, run PrepSCTFindMarkers and
generate markers for the alternative clustering results. Bonus to
generate a table of the top 5 markers and outputing that to file.
Check-in Question - clustering impact on marker genes After you generate markers for the “fewer”
pcsoption, how do the results differ from the markers found forpcs=10in the workshop session? What do you think would happen if you tried this with the “more” results?
Day 3 Exercise 2 - Generate predictions for alternative clustering
Using scCATCH like we did for our annotation process, generate predictions for the alternative clustering results.
Day 3 Exercise 3 - Add predictions to Seurat object
Skipping the refinement step, add the cell-type predictions to the Seurat object using the numeric labels as a key, similarly to the our annotation step.
Day 3 Exercise 4 - Plot UMAP with new cluster labels
Using a similar approach used to visualize the annotations in the workshop, generate a UMAP plot
Hint - use the same reduction name as yesterday’s exercise
and make sure the group_by parameter matches the column
name added to the metadata that stores the predicted cell-types.
Check-in Question
How did the number of pcs and/or resolution change the predictions? Do you think the predictions correspond better or worse to the cluster structure we see in the UMAP?
Clean up session
Before closing out the window, make sure to clear environment and restart R session to manage the memory usage
## ----------------------------------------------------------
## Clean up session
rm(list=names(which(unlist(eapply(.GlobalEnv, is.ggplot)))));
rm(catch_celltypes, catch_markers, geo2_catch, geo2_markers, new_metadata, top_5);
gc()
## (Optional) - Save copy of exso3
saveRDS(exso3, file = paste0('results/rdata/geo_so_sct_integrated_with_markers_exercise.rds'))
## BEFORE CLOSING WINDOW - POWER DOWN AND RESTART R SESSION
Day 3 solutions
Scripts
# =========================================================================
# Independent Exercise - Day 3 Startup
# =========================================================================
# After restarting our R session, load the required libraries
library(Seurat)
library(BPCells)
library(tidyverse)
library(scCATCH)
# Load in seurat object with alternative clustering results from yesterday's exercises
exso3 = readRDS('results/rdata/geo_so_sct_clustered_exercise.rds')
exso3 # check that object loaded
## NOTE - BEFORE STOPPING WORK ON THE EXERCISES REMEMBER TO POWER DOWN AND RESTART R SESSION !!!!
### Day 3 Exercise 1 - examine marker genes and cell type predictions for alternative clustering
## ----------------------------------------------------------
# Check what identities are set
Idents(exso3) %>% head()
## use same values as previous exercise
pcs = 6
res = 0.2
## Set identities to clustering for selected resolution
exso3 = SetIdent(exso3, value = paste0('int.sct.rpca.clusters', res))
Idents(exso3) %>% head()
## ----------------------------------------------------------
## Generate cluster markers to see how that changes with new parameters
exso3 = PrepSCTFindMarkers(exso3, assay = "SCT") # NOTE - this step will take some time to run
exso3_markers = FindAllMarkers(exso3, only.pos = TRUE)
head(exso3)
# Create table of top 5 markers per cluster (using default ranking)
top_5 = exso3_markers %>% filter(p_val_adj < 0.01) %>% group_by(cluster) %>% slice_head(n = 5)
head(top_5, n = 10) # look at results
write_csv(top_5, file = paste0('results/tables/top5_marker_genes_exercise.csv'))
#### Day 3 Exercise 2 - Generate predictions for alternative clustering
## ----------------------------------------------------
## Next - run scCATCH predictions for alternative clustering results
exso3_catch = createscCATCH(data = exso3@assays$SCT@counts, cluster = as.character(Idents(exso3)))
catch_markers = exso3_markers %>% rename('logfc' = 'avg_log2FC')
exso3_catch@markergene = exso3_markers
exso3_catch@marker = cellmatch[cellmatch$species == 'Mouse' & cellmatch$tissue %in% c('Blood', 'Peripheral Blood', 'Muscle', 'Skeletal muscle', 'Epidermis', 'Skin'), ]
exso3_catch = findcelltype(exso3_catch)
# Check predictions
exso3_catch@celltype %>% select(cluster, cell_type, celltype_score)
#### Day 3 Exercise 3 - Add predictions to Seurat object
## ------------------------------------------------------
## Use predictions to label clusters and UMAP plot
catch_celltypes = exso3_catch@celltype %>% select(cluster, cell_type)
colnames(catch_celltypes)[2] = paste0('cell_type.',pcs,'PC.',res,'res')
new_metadata = exso3@meta.data %>%
left_join(catch_celltypes,
by = c('seurat_clusters' = 'cluster')) # using `seurat_clusters`, which will store the most recently generated cluster labels for each cell
rownames(new_metadata) = rownames(exso3@meta.data) # We are implicitly relying on the same row order!
exso3@meta.data = new_metadata # Replace the meta.data
head(exso3@meta.data)
#### Day 3 Exercise 4 - Plot UMAP with new cluster labels
## ------------------------------------------------------
catch_umap_plot = DimPlot(exso3, group.by = paste0('cell_type.',pcs,'PC.',res,'res'),
label = TRUE, reduction = paste0('umap.integrated.sct.rpca_alt', res))
catch_umap_plot
# Save the plot to file
# output to file, including the number of PCs and resolution used to generate results
ggsave(filename = paste0('results/figures/umap_int_catch-labeled_',
pcs,'PC.',res,'res','.png'),
plot = catch_umap_plot,
width = 8, height = 6, units = 'in')
## ----------------------------------------------------------
## Clean up session
rm(list=names(which(unlist(eapply(.GlobalEnv, is.ggplot)))));
rm(catch_celltypes, catch_markers, exso3_catch, exso3_markers, new_metadata, top_5);
gc()
## (Optional) - Save copy of exso3
saveRDS(exso3, file = paste0('results/rdata/geo_so_sct_integrated_with_markers_exercise.rds'))
rm(exso3)
gc()
## BEFORE PROCEEDING TO THE NEXT SECTION or closing window - POWER DOWN AND RESTART R SESSIONFull output
# =========================================================================
# Independent Exercise - Day 3 Startup
# =========================================================================
# After restarting our R session, load the required libraries
library(Seurat)
library(BPCells)
library(tidyverse)
library(scCATCH)
# Load in seurat object with alternative clustering results from yesterday's exercises
exso3 = readRDS('results/rdata/geo_so_sct_clustered_exercise.rds')
exso3 # check that object loadedAn object of class Seurat
46957 features across 31559 samples within 2 assays
Active assay: SCT (20468 features, 3000 variable features)
3 layers present: counts, data, scale.data
1 other assay present: RNA
6 dimensional reductions calculated: unintegrated.sct.pca, integrated.sct.rpca, umap.integrated.sct.rpca_alt0.2, umap.integrated.sct.rpca_alt0.4, umap.integrated.sct.rpca_alt0.8, umap.integrated.sct.rpca_alt1## NOTE - BEFORE STOPPING WORK ON THE EXERCISES REMEMBER TO POWER DOWN AND RESTART R SESSION !!!!
### Day 3 Exercise 1 - examine marker genes and cell type predictions for alternative clustering
## ----------------------------------------------------------
# Check what identities are set
Idents(exso3) %>% head()HODay0replicate1_AAACCTGAGAGAACAG-1 HODay0replicate1_AAACCTGGTCATGCAT-1 HODay0replicate1_AAACCTGTCAGAGCTT-1 HODay0replicate1_AAACGGGAGAGACTTA-1 HODay0replicate1_AAACGGGAGGCCCGTT-1
3 3 1 23 9
HODay0replicate1_AAACGGGCAACTGGCC-1
3
Levels: 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29## use same values as previous exercise
pcs = 6
res = 0.2
## Set identities to clustering for selected resolution
exso3 = SetIdent(exso3, value = paste0('int.sct.rpca.clusters', res))
Idents(exso3) %>% head()HODay0replicate1_AAACCTGAGAGAACAG-1 HODay0replicate1_AAACCTGGTCATGCAT-1 HODay0replicate1_AAACCTGTCAGAGCTT-1 HODay0replicate1_AAACGGGAGAGACTTA-1 HODay0replicate1_AAACGGGAGGCCCGTT-1
0 0 0 7 0
HODay0replicate1_AAACGGGCAACTGGCC-1
0
Levels: 0 1 2 3 4 5 6 7 8 9 10 11## ----------------------------------------------------------
## Generate cluster markers to see how that changes with new parameters
exso3 = PrepSCTFindMarkers(exso3, assay = "SCT") # NOTE - this step will take some time to runFound 12 SCT models. Recorrecting SCT counts using minimum median counts: 3675.5exso3_markers = FindAllMarkers(exso3, only.pos = TRUE)Calculating cluster 0Calculating cluster 1Calculating cluster 2Calculating cluster 3Calculating cluster 4Calculating cluster 5Calculating cluster 6Calculating cluster 7Calculating cluster 8Calculating cluster 9Calculating cluster 10Calculating cluster 11head(exso3) orig.ident nCount_RNA nFeature_RNA condition time replicate percent.mt nCount_SCT nFeature_SCT int.sct.rpca.clusters0.2 seurat_clusters
HODay0replicate1_AAACCTGAGAGAACAG-1 HODay0replicate1 10234 3226 HO Day0 replicate1 1.240962 6062 2867 0 3
HODay0replicate1_AAACCTGGTCATGCAT-1 HODay0replicate1 3158 1499 HO Day0 replicate1 7.536415 4607 1509 0 3
HODay0replicate1_AAACCTGTCAGAGCTT-1 HODay0replicate1 13464 4102 HO Day0 replicate1 3.112002 5314 2370 0 1
HODay0replicate1_AAACGGGAGAGACTTA-1 HODay0replicate1 577 346 HO Day0 replicate1 1.559792 3877 1031 7 23
HODay0replicate1_AAACGGGAGGCCCGTT-1 HODay0replicate1 1189 629 HO Day0 replicate1 3.700589 4166 915 0 9
HODay0replicate1_AAACGGGCAACTGGCC-1 HODay0replicate1 7726 2602 HO Day0 replicate1 2.938131 5865 2588 0 3
HODay0replicate1_AAACGGGGTCCGAATT-1 HODay0replicate1 5165 2362 HO Day0 replicate1 9.196515 5162 2348 6 11
HODay0replicate1_AAACGGGGTCGGATCC-1 HODay0replicate1 2379 1381 HO Day0 replicate1 2.564103 4682 1400 6 11
HODay0replicate1_AAACGGGTCAGCTGGC-1 HODay0replicate1 12160 3646 HO Day0 replicate1 2.023026 5765 2604 0 8
HODay0replicate1_AAACGGGTCCGTACAA-1 HODay0replicate1 9864 2800 HO Day0 replicate1 2.139092 6033 2570 0 3
int.sct.rpca.clusters0.4 int.sct.rpca.clusters0.8 int.sct.rpca.clusters1
HODay0replicate1_AAACCTGAGAGAACAG-1 4 5 3
HODay0replicate1_AAACCTGGTCATGCAT-1 4 5 3
HODay0replicate1_AAACCTGTCAGAGCTT-1 0 1 1
HODay0replicate1_AAACGGGAGAGACTTA-1 17 13 23
HODay0replicate1_AAACGGGAGGCCCGTT-1 1 0 9
HODay0replicate1_AAACGGGCAACTGGCC-1 4 1 3
HODay0replicate1_AAACGGGGTCCGAATT-1 8 11 11
HODay0replicate1_AAACGGGGTCGGATCC-1 8 11 11
HODay0replicate1_AAACGGGTCAGCTGGC-1 1 0 8
HODay0replicate1_AAACGGGTCCGTACAA-1 4 5 3# Create table of top 5 markers per cluster (using default ranking)
top_5 = exso3_markers %>% filter(p_val_adj < 0.01) %>% group_by(cluster) %>% slice_head(n = 5)
head(top_5, n = 10) # look at results# A tibble: 10 × 7
# Groups: cluster [2]
p_val avg_log2FC pct.1 pct.2 p_val_adj cluster gene
<dbl> <dbl> <dbl> <dbl> <dbl> <fct> <chr>
1 0 5.36 0.826 0.066 0 0 Clec3b
2 0 3.37 0.883 0.159 0 0 Dpt
3 0 3.39 0.957 0.244 0 0 Serping1
4 0 3.63 0.778 0.117 0 0 Col14a1
5 0 3.27 0.757 0.103 0 0 C1s1
6 0 4.22 0.921 0.135 0 1 Col12a1
7 0 4.09 0.833 0.1 0 1 Tnc
8 0 3.75 0.885 0.156 0 1 Thbs2
9 0 2.99 0.894 0.169 0 1 Lox
10 0 4.97 0.756 0.064 0 1 Col11a1 write_csv(top_5, file = paste0('results/tables/top5_marker_genes_exercise.csv'))
#### Day 3 Exercise 2 - Generate predictions for alternative clustering
## ----------------------------------------------------
## Next - run scCATCH predictions for alternative clustering results
exso3_catch = createscCATCH(data = exso3@assays$SCT@counts, cluster = as.character(Idents(exso3)))
catch_markers = exso3_markers %>% rename('logfc' = 'avg_log2FC')
exso3_catch@markergene = exso3_markers
exso3_catch@marker = cellmatch[cellmatch$species == 'Mouse' & cellmatch$tissue %in% c('Blood', 'Peripheral Blood', 'Muscle', 'Skeletal muscle', 'Epidermis', 'Skin'), ]
exso3_catch = findcelltype(exso3_catch)[+++++++>----------------------] Finished: 25% Time 00:00:00[+++++++++>--------------------] Finished: 33% Time 00:00:00[+++++++++++>------------------] Finished: 42% Time 00:00:00[++++++++++++++>---------------] Finished: 50% Time 00:00:00[+++++++++++++++++>------------] Finished: 58% Time 00:00:01[+++++++++++++++++++>----------] Finished: 67% Time 00:00:01[+++++++++++++++++++++>--------] Finished: 75% Time 00:00:01[++++++++++++++++++++++++>-----] Finished: 83% Time 00:00:01[+++++++++++++++++++++++++++>--] Finished: 92% Time 00:00:02[++++++++++++++++++++++++++++++] Finished:100% Time 00:00:02# Check predictions
exso3_catch@celltype %>% select(cluster, cell_type, celltype_score) cluster cell_type celltype_score
1 0 Hematopoietic Stem Cell 0.87
2 1 Stem Cell 0.83
3 2 Macrophage 0.83
4 3 Neutrophil 0.81
5 4 Dendritic Cell 0.86
6 5 Germinal Center B Cell 0.88
7 6 Hematopoietic Stem Cell 0.91
8 7 Hematopoietic Stem Cell 0.90
9 8 Hematopoietic Stem Cell 0.88
10 9 Muscle Satellite Cell 0.95
11 10 CD8+ T Cell 0.90
12 11 Stromal Cell 0.66#### Day 3 Exercise 3 - Add predictions to Seurat object
## ------------------------------------------------------
## Use predictions to label clusters and UMAP plot
catch_celltypes = exso3_catch@celltype %>% select(cluster, cell_type)
colnames(catch_celltypes)[2] = paste0('cell_type.',pcs,'PC.',res,'res')
new_metadata = exso3@meta.data %>%
left_join(catch_celltypes,
by = c('seurat_clusters' = 'cluster')) # using `seurat_clusters`, which will store the most recently generated cluster labels for each cell
rownames(new_metadata) = rownames(exso3@meta.data) # We are implicitly relying on the same row order!
exso3@meta.data = new_metadata # Replace the meta.data
head(exso3@meta.data) orig.ident nCount_RNA nFeature_RNA condition time replicate percent.mt nCount_SCT nFeature_SCT int.sct.rpca.clusters0.2 seurat_clusters
HODay0replicate1_AAACCTGAGAGAACAG-1 HODay0replicate1 10234 3226 HO Day0 replicate1 1.240962 6062 2867 0 3
HODay0replicate1_AAACCTGGTCATGCAT-1 HODay0replicate1 3158 1499 HO Day0 replicate1 7.536415 4607 1509 0 3
HODay0replicate1_AAACCTGTCAGAGCTT-1 HODay0replicate1 13464 4102 HO Day0 replicate1 3.112002 5314 2370 0 1
HODay0replicate1_AAACGGGAGAGACTTA-1 HODay0replicate1 577 346 HO Day0 replicate1 1.559792 3877 1031 7 23
HODay0replicate1_AAACGGGAGGCCCGTT-1 HODay0replicate1 1189 629 HO Day0 replicate1 3.700589 4166 915 0 9
HODay0replicate1_AAACGGGCAACTGGCC-1 HODay0replicate1 7726 2602 HO Day0 replicate1 2.938131 5865 2588 0 3
int.sct.rpca.clusters0.4 int.sct.rpca.clusters0.8 int.sct.rpca.clusters1 cell_type.6PC.0.2res
HODay0replicate1_AAACCTGAGAGAACAG-1 4 5 3 Neutrophil
HODay0replicate1_AAACCTGGTCATGCAT-1 4 5 3 Neutrophil
HODay0replicate1_AAACCTGTCAGAGCTT-1 0 1 1 Stem Cell
HODay0replicate1_AAACGGGAGAGACTTA-1 17 13 23 <NA>
HODay0replicate1_AAACGGGAGGCCCGTT-1 1 0 9 Muscle Satellite Cell
HODay0replicate1_AAACGGGCAACTGGCC-1 4 1 3 Neutrophil#### Day 3 Exercise 4 - Plot UMAP with new cluster labels
## ------------------------------------------------------
catch_umap_plot = DimPlot(exso3, group.by = paste0('cell_type.',pcs,'PC.',res,'res'),
label = TRUE, reduction = paste0('umap.integrated.sct.rpca_alt', res))
catch_umap_plot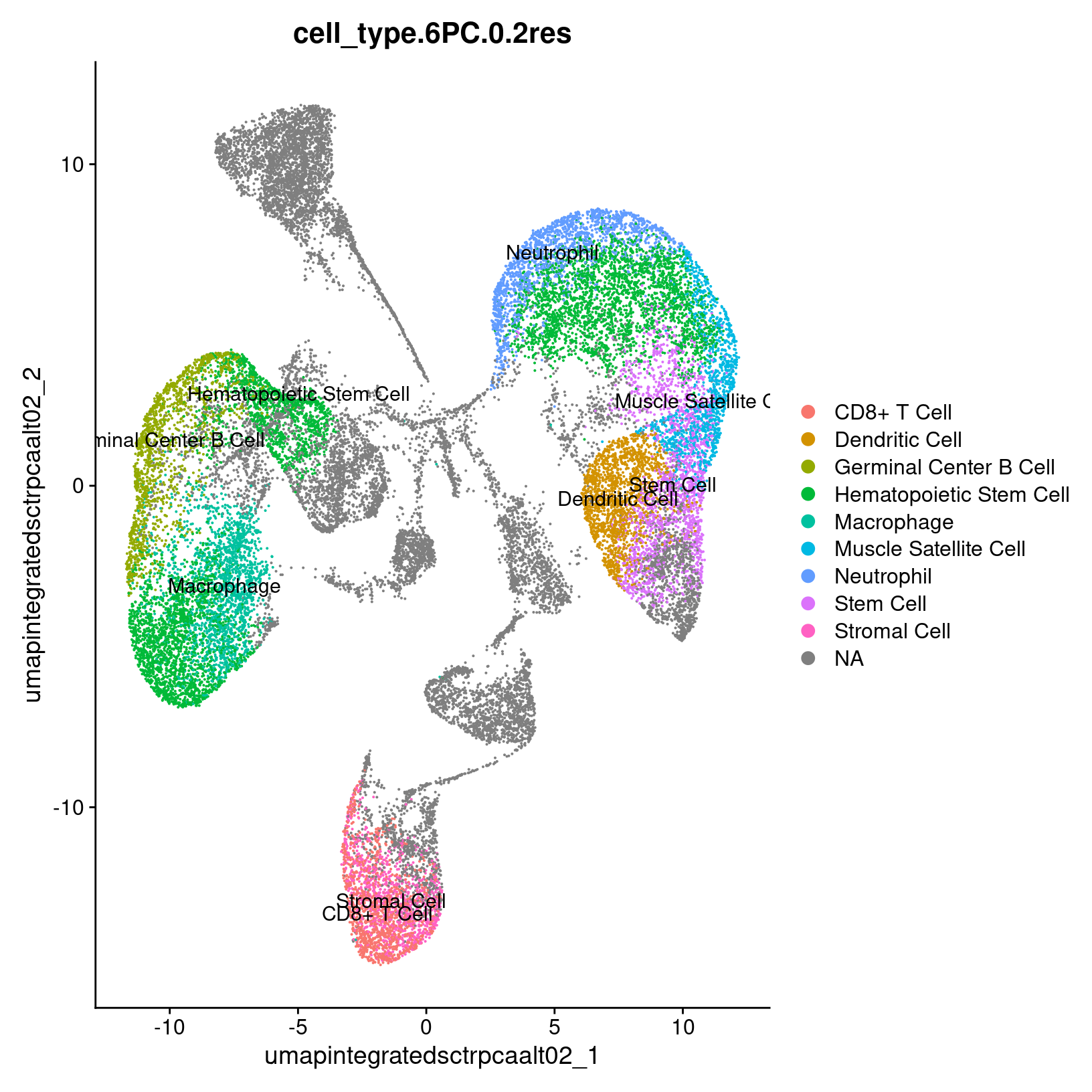
# Save the plot to file
# output to file, including the number of PCs and resolution used to generate results
ggsave(filename = paste0('results/figures/umap_int_catch-labeled_',
pcs,'PC.',res,'res','.png'),
plot = catch_umap_plot,
width = 8, height = 6, units = 'in')
## ----------------------------------------------------------
## Clean up session
rm(list=names(which(unlist(eapply(.GlobalEnv, is.ggplot)))));
rm(catch_celltypes, catch_markers, exso3_catch, exso3_markers, new_metadata, top_5);
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 6944763 370.9 10752860 574.3 10752860 574.3
Vcells 338468682 2582.4 1229359917 9379.3 1534788602 11709.6## (Optional) - Save copy of exso3
saveRDS(exso3, file = paste0('results/rdata/geo_so_sct_integrated_with_markers_exercise.rds'))
rm(exso3)
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 6944784 370.9 10752860 574.3 10752860 574.3
Vcells 338468716 2582.4 983487934 7503.5 1534788602 11709.6## BEFORE PROCEEDING TO THE NEXT SECTION or closing window - POWER DOWN AND RESTART R SESSION